PROJECT DESCRIPTION
A TOOL FOR DIAGNOSING IBD BASED ON ASSOCIATIVE LEARNING
The ETH Zurich iGEM team 2016 will focus on the diagnostic monitoring of inflammatory bowel disease. Current diagnostic methods are invasive and rely on biomarkers that are not sufficiently disease-specific. We have engineered Escherichia coli that have the potential to detect several disease-specific biomarkers and to memorize this event. While Pavlov's Coli travels through the gut, simultaneous detection of IBD markers activate an AND gate which triggers a unidirectional switch, thus committing the observation to memory. The addition of a candidate biomarker to the bacteria isolated from the patient's faeces induces the expression of a fluorescent protein, if this biomarker has been memorized. Thus, a single reporter is sufficient to differentiate between many different candidate markers. A community of sensor cells with different sensing specificities can be utilized at the same time, enabling a high degree of multiplexing. Pavlov’s Coli holds promise as a non-invasive diagnostic tool, potentially able to shed light on microbiome imbalances associated with IBD.
INFLAMMATORY BOWEL DISEASE
Inflammatory bowel disease (IBD) is characterised by chronic inflammation of parts of the intestine. The term describes an ensemble of conditions, the most common of which are ulcerative colitis and Crohn's disease. IBD is classified as an autoimmune disease for which no cure has been developed. Current treatments include immunosuppression, surgery, antibiotics and nutritional therapies. IBD is not only a severe burden for the patients, but it also causes increasing direct and indirect costs for society. Furthermore, only in Europe 2.5 million people are affected by IBD and the number of cases is increasing world-wide1.
Unfortunately there are no characteristic blood markers to distinguish between the different forms of IBD. The diagnosis is mostly based on the location of inflammation observed during a colonoscopy. Also the underlying trigger of the disease is not completely understood but correlation studies proposed factors such as diet, genetic predisposition, breach of the intestinal barrier and unfavorable alteration of the microbiota, called dysbiosis2. The diversity of the microbiota is noticeably reduced3,4 in IBD patients and the composition of the gut flora changes from symbiotic to predominantly pathobiotic microbes5.
Inflammation partially disrupts the integrity of the layer of epithelial cells lining the intestine. This cell layer separates the gut lumen containing trillions of microbes from the body. The damage to this essential barrier compromises its selectivity and allows for penetration of immunogenic antigens from the lumen across the epithelial layer6,7 which enhances the inflammation reaction.
SENSING OF MARKERS FOR IBD
NITRIC OXIDE

Figure 1: NorR constitutively binds to DNA. Only when NO• is present it activates the transcription of the gene under control of the norVW promoter (adapted from Green et al.)
Beside the penetration of immunogenic antigens across the epithelial layer, there is also abnormal leakage of inflammation markers into the gut lumen. One such molecule is nitric oxide (NO•, t1/2 < 6 seconds8). Since IBD is normally accompanied by severe inflammation, we chose to develop a sensor for NO. The sensing of NO• with E. coli has already been described by Archer et al.9 in 2012. This work provides us with the relevant genetic elements and helps us to design a sensor for our purpose. Additionally, the authors present their system as a rapid detection strategy for diagnosing IBD related disease flare-ups, which would allow for immediate intervention.
NorR is capable of binding NO• via its mononuclear non-heme iron center. While other sensor proteins are not only specific for NO• but also for other NOx species, NorR specifically binds the NO• radical. Hexameric NorR is constitutively bound upstream of the norVW promoter, but in the absence of NO• its AAA+ domain is blocked. ATP hydrolysis by the AAA+ domain of NorR provides the necessary energy for the open complex formation involving the DNA and the RNA-polymerase holoenzyme. In the absence of NO•, this domain is blocked and transcription initiation is prevented. Upon binding of NO• a conformational change in NorR frees the AAA+ domain which allows for transcription initiation.10. Based on these considerations, we included in our detection system the promoter element 'PnorV', which is resposive to NO-activated norR, allowing us to sense the presence of nitric oxide.
N-ACYL HOMOSERINE LACTONES

Figure 2: DNA- and 3-oxo-C6-HSL bound, dimeric form of TraR, a close homolog of EsaR (PDB: 1L3L, edited with UCSF Chimera11)
Our goal is to not only detect a general inflammation biomarker, but also specific molecules secreted by the microbiota, which would allow us to gain insights into microbiome composition. One well-known class of molecules secreted by many bacterial species belongs to the quorum sensing (QS) system. QS molecules act as bacterial hormones allowing intra- and interspecies communication. Among other things, they control biofilm formation and growth behaviour. Furthermore, manipulation of QS signals has been found to affect the gut microbiota and its composition12. The best known subclass of QS molecules are the N-acyl homoserine lactones (AHL). Previous findings have shown abnormal levels of a number of AHL species in IBD patients. One of the AHLs that has been found to be upregulated in IBD is 3-hydroxy-hexanoyl-HSL (3-OH-C6-HSL)13. A well characterized regulatory protein that senses a very similar HSL (3-oxo-C6-HSL) is EsaR from Erwinia stewartii that was used by a former iGEM team. The special feature of EsaR is its regulatory behaviour: while most HSL-responsive elements are inducible activators, EsaR is a repressor that dissociates from the DNA in the presence of HSL. This is important when designing a genetic circuit as a repressor is thought to be less leaky than an activator. Based on all these considerations, we chose HSL (3-oxo-C6-HSL) as the microbiome-associated target to be detected by our genetically engineered E. coli. As our target HSL is not the natural ligand for EsaR, we designed and commenced experiments for the directed evolution of EsaR in order to change its specificity.
ASSOCIATIVE LEARNING CIRCUIT
OVERVIEW
In order to serve as a diagnostics and research tool, our system should not only be able to sense a single molecule alone but should associate an inflammation marker - in our case NO• - with a potential trigger of the inflammation itself. Thus, we implemented an associative learning circuit that allows for the detection of the temporal and spatial presence of two markers. (See Design)
Nitric oxide and 3-OH-C6-HSL are only two possible markers of IBD. There are many more potential biomarkers that are definitively worth investigating. To account for this multitude of possible markers, we devised a modular system that allows for rapid component exchange. Moreover, we extended our single AND-gate by the addition of a memory module: AND-gate activation results in the activation of a unidirectinal switching mechanism, which in turn renders a reporter protein (e.g. GFP) sensitive to a bio-marker previuosly encountered by the AND-gate (e.g. AHL), which causes reporter expression. While the number of distinguistable reporters (e.g. fluorophores) is limited, our system allows for simultaneous observation of a multitude of markers measured in parallel. Our Pavlov's Coli learn the occurence of the presence of two markers and store this information in their DNA until readout.
We designed our system in a way that allows fast and easy demultiplexing of a complex mixture of different reporter strains. If the reporter strains encounter the inflammation-associated marker again, they generate an easily observable output: fluorescence. This was achieved by integrating a second AND-gate that relies on the successful learning process as shown below.
SENSOR AND-GATE:
At an inflammation spot, nitric oxide activates NorR and triggers the transcription of the bxb1 integrase gene. Yet, the transcription can proceed only if OH-C6-HSL is also present. HSL makes the repressor EsaR dissociate from the regulatory element (esaBox) on the DNA and thus annihiliates its roadblock activity.
LEARNING:
Once the bxb1 gene is successfully transcribed and translated, Bxb1 binds to the attP and attB recombination sites flanking a constitutive promoter and inverts it. As attP and attB are destroyed through inversion, Bxb1 mediated recombination acts as a one-way switch.
REPORTER AND-GATE:
The constitutive promoter, which is now placed upstream of the reporter protein GFP, is additionally under the control of another esaBox, the binding site of EsaR.
Now that the system has learned to respond to the associated stimulus, namely the AHL alone, the expression of GFP is induced whenever the stimulus, i.e. EsaR's ligand, is encountered again.
BIOLOGICAL IMPLEMENTATION: CRISPR/Cpf1
An alternative to a recombinase-based switch is the usage of the 2015 characterised CRISPR/Cpf1 system14. Instead of cutting both DNA strands at the same position, Cpf1 cuts the DNA with an offset of four or five nucleotides, thus producing single-stranded overhangs. It is suggested that this is advantageous for genome editing via non-homologous end-joining15.
Cpf1 is potentially capable of creating an AND-gate controlled one-way switch with finally the same functionality as the recombinase-based switch.
For this, a reporter construct needs to be stably integrated into the genome of E. coli whereas Cpf1 and its guide RNAs are expressed from a plasmid.
SENSOR AND-GATE:
At a site of inflammation, nitric oxide activates NorR and triggers the transcription of Cfp1. The transcription can only proceed if 3-OH-C6-HSL is present. The HSL makes the repressor EsaR dissociate from the regulatory element (esaBox) on the DNA and thus annihiliates its roadblock activity. The guide RNAs are expressed constitutively at a high level.
LEARNING:
Once Cfp1 is expressed, it is brought to the cleavage-sites by the two distinct guide RNAs. There, Cfp1 cuts out the mNectarine gene while creating sticky ends. These will then be ligated by endogeneous ligases via NHEJ which reconstitutes the GFP gene.
REPORTER AND-GATE:
In a final system, the gfp gene would be under the control of a constitutive promoter regulated by an esaBox, the binding site of EsaR.
After the system has now learned to respond to the associated stimulus alone, the expression of GFP could easily be induced by just exposing it to the stimulus again, e.g. EsaR's ligand.
BIOLOGICAL IMPLEMENTATION: CRISPR/Cas9
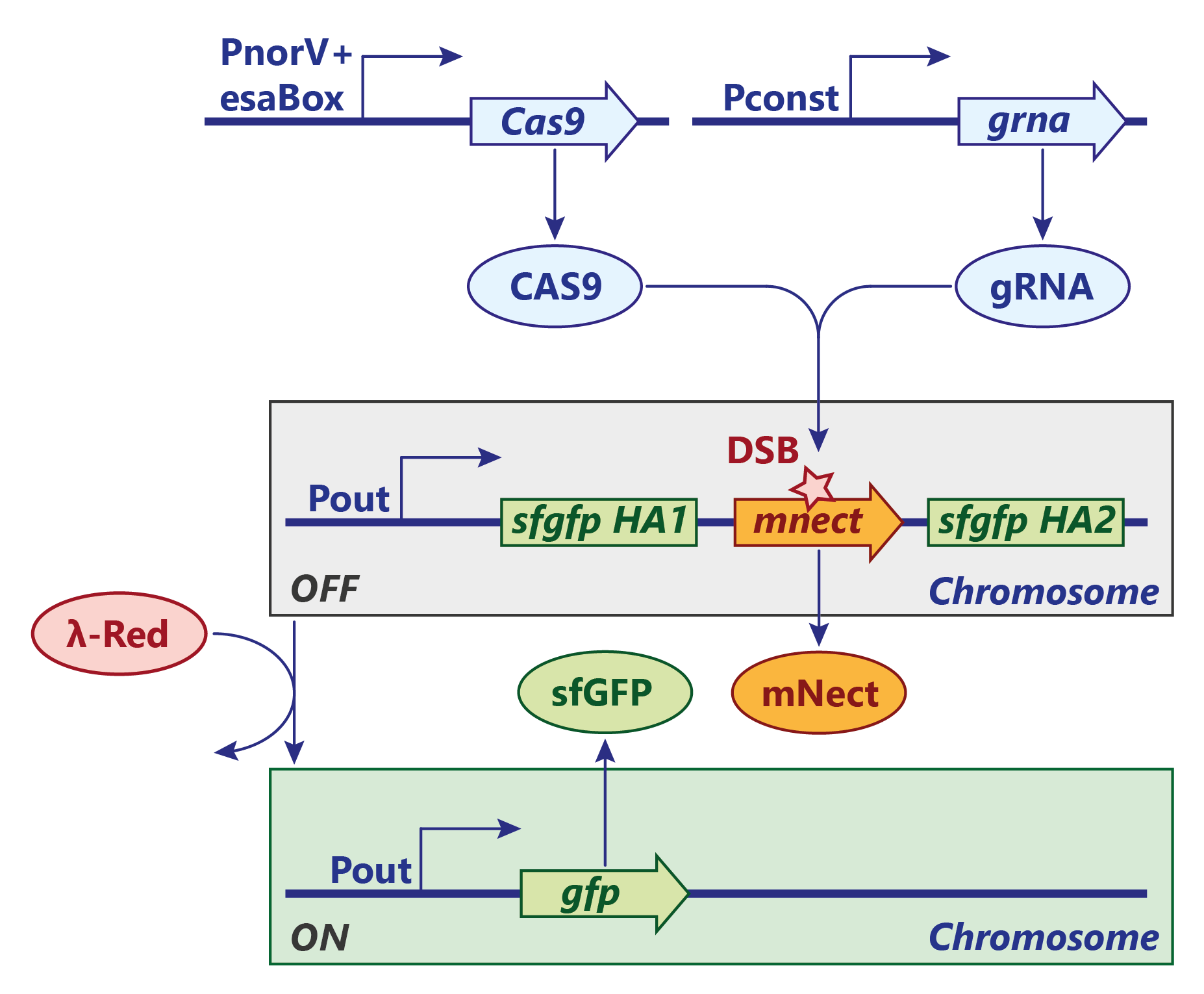
A similar approach for an AND gate with an irreverisble genetic switch was persued using the classic CRISPR/Cas9 system. Contrary to the CRISPR/Cpf1 system, the Cas9 endonuclease cuts the genome integrated reporter construct only once and the double strand breake (DSB) is sealed by homology directed repair. In our test system, the λ-Red system, which is necessary for recombination, is induced by a heat shock but would finally be under the same regulation as Cas9. Furthermore, the expression of mNectarine / GFP would be under control of the candidate molecule (e.g. 3-OH-C6-HSL).
SENSOR AND-GATE:
At a site of inflammation, nitric oxide activates NorR and triggers the transcription of Cas9. The transcription can only proceed if 3-OH-C6-HSL is present. The HSL makes the repressor EsaR dissociate from the regulatory element (esaBox) on the DNA and thus annihiliates its roadblock activity. The guide RNA is expressed constitutively at a high level.
LEARNING:
Once Cas9 is expressed, it is brought to the cleavage-site by the the guide RNA. There, Cas9 cuts the mNectarine gene while introducing a double strand break. The repair mechanism by the λ-Red system is - in our test system - manually induced by a heat shock which then recombines the gfp gene via the two homology arms. This way, mNectarine is removed and GFP is expressed.
REPORTER AND-GATE:
In a final system, the gfp gene would be under the control of a constitutive promoter regulated by an esaBox, the binding site of EsaR.
After the system has now learned to respond to the associated stimulus alone, the expression of GFP could easily be induced by just exposing it to the stimulus again, e.g. EsaR's ligand.
DIRECTED EVOLUTION OF EsaR
In order to change EsaR's specificity towards an IBD related HSL, we try to to apply directed evolution on this repressor protein.
The variant of EsaR that we used was already an improved version with a D91G mutation that has an increased signal sensitivity compared to the wildtype16.A dual selection system offers a possibility to select for these variants. These systems allow for negative selection ("killing") of variants that still react to the native HSL (3-oxo-C6-HSL) and positive selection ("survival") of variants that respond to the new target HSL (3-OH-C6-HSL). We used a fusion protein that is composed of a domain which confers antibiotic resistance and an enzyme that converts a non-toxic compound into a cellular toxin. We tested the combination of the chloramphenicol acetyltransferase (CAT) and the uracil phosphoribosyltransferase (UPRT)17. While CAT confers resistance for the positive selection step, UPRT is necessary for the negative selection.
UPRT normally converts uracil into uridine monophosphate (dUMP). 5-fluorouracil is metabolised by UPRT to 5-fluoro-dUMP. This irreversibly blocks the thymidylate synthase (thyA), a key enzyme in the production of pyrimidine nucleosides in the cell, which finally leads to cell death18.
DUAL SELECTION PROCEDURE
NEGATIVE SELECTION:
In a first step, the created library of variants is grown in the presence of the old inducer 3-oxo-C6-HSL and a toxin precursor.
Variants whose expression is still induced by the old HSL or which have a non-functional repressor (EsaR) express the fusion protein which converts the toxin precursor into a toxin (A), whereas non-responsive repressors remain bound to the DNA and inhibit protein expression (B).
POSITIVE SELECTION:
In a second step, these variants now undergo a round of positive selection to select for variants that are responsive to the new HSL 3-OH-C6-HSL.
The bacteria are cultured in medium containing the new HSL and the antibiotic whose resistance is part of the fusionprotein. Inducible variants express the resistance protein and survive (A). Variants that are non-responsive do not express it and can not grow (B).
Afterwards, the variants can be plated and analyzed or subjected to further rounds of positive / negative selection to enrich for suitable variants.
POTENTIAL DELIVERY METHOD OF REPORTER STRAINS
The idea to administer genetically modified bacteria in the context of IBD was put forward by Steidler et al. in 200019. Generally, the use of engineered probiotic bacteria as a delivery vector for in vivo produced therapeutic agents has been described multiple times.As the gastrointestinal tract is a rough environment for non-adapted (probiotic) bacteria, we envision our bacteria to be encapsulated in a hydrogel-coated pill. This would protect the bacteria and ensure the recovery of the reporter strain. The method of encapsulation is well known for oral administration in animal models (e.g. Prakash et al.20) and is summarised in several reviews.
READOUT OF RECOVERED REPORTER BACTERIA
Our complex design with its potentially modular setup serves mainly one goal: to measure multiple candidate molecules in parallel through several bacterial reporter strains combined in one pill. Thus, it is curcial to understand how the read-out of our system works, once the reporter strains are recovered from a patient.
The recovered bacterial strains now form a complex mixture and it is not possible to separate them again. But with our system this is not even necessary. The procedure to easily figure out if a specific candidate molecule has been associated with inflammation is the following:
- The recovered bacteria-containing capsules are split into different vials. Each vial thus contains the same composition of bacterial reporter strains.
- To each vial a different candidate molecule is added.
-
In case one of the bacterial strains has encountered this specific molecule it will "remember" (as their plasmids / genomes made an irreversible rearrangement due to recombinases, Cpf1 or Cas9).
- Cultures which show an a green colour (GFP, positive signal) - additionally to orange (mNectarine, negative control) - have successfully associated intestinal inflammation with the candidate molecule.
-
mNectarine will always be present, as only one or no strain will react to the candidate molecule.
IMPROVED PARTS
The parts we used for our constructs were partially already described and used by former iGEM teams. We have three major parts that we could improve - either be codon optimization and/or characterization. These are BxB1 recombinase, tp901 and the pNorV promoter. Please see our Demonstrate for further information.
REFERENCES:
- [1] Kaplan, Gilaad G. "The global burden of IBD: from 2015 to 2025." Nature Reviews Gastroenterology & Hepatology 12.12 (2015): 720-727.
- [2] Scaldaferri, F., et al. "Gut microbiota molecular spectrum in healthy controls, diverticular disease, IBS and IBD patients: Time for microbial marker of gastrointestinal disorders?." Journal of Crohns & Colitis. Vol. 9. Oxford Univ Press, 2015.
- [3] Ott, S. J., and S. Schreiber. "Reduced microbial diversity in inflammatory bowel diseases." Gut 55.8 (2006): 1207-1207.
- [4] Hold, Georgina L., et al. "Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years." World J Gastroenterol 20.5 (2014): 1192-1210.
- [5] Kaur, Nirmal, et al. "Intestinal dysbiosis in inflammatory bowel disease." Gut microbes 2.4 (2011): 211-216.
- [6] Chichlowski, Maciej, and Laura P. Hale. "Bacterial-mucosal interactions in inflammatory bowel disease—an alliance gone bad." American Journal of Physiology-Gastrointestinal and Liver Physiology 295.6 (2008): G1139-G1149.
- [7] Laukoetter, Mike G., Porfirio Nava, and Asma Nusrat. "Role of the intestinal barrier in inflammatory bowel disease." World Journal of Gastroenterology 14.3 (2008): 401.
- [8] Kochar, Nitin I., et al. "Nitric oxide and the gastrointestinal tract." Int. J. Pharmacol 7 (2011): 31-39.
- [9] Archer, Eric J., Andra B. Robinson, and Gürol M. Süel. "Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing." ACS synthetic biology 1.10 (2012): 451-457.
- [10] Green, Jeffrey, Matthew D. Rolfe, and Laura J. Smith. "Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide." Virulence 5.8 (2014): 794-809.
- [11] UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
- [12] Thompson, Jessica Ann, et al. "Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota." Cell reports 10.11 (2015): 1861-1871.
- [13] Landman, Cecilia, et al. "Sa1804 Quorum Sensing Driven by N-Acyl-Homoserine Lactone in Inflammatory Bowel Diseases Associated Dysbiosis." Gastroenterology 144.5 (2013): S-310.
- [14] Zetsche, Bernd, et al. "Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system." Cell 163.3 (2015): 759-771.
- [15] Fagerlund, Robert D., Raymond HJ Staals, and Peter C. Fineran. "The Cpf1 CRISPR-Cas protein expands genome-editing tools." Genome biology 16.1 (2015): 1.
- [16] Shong, Jasmine, et al. "Directed evolution of the quorum-sensing regulator EsaR for increased signal sensitivity." ACS chemical biology 8.4 (2013): 789-795.
- [17] Rackham, Oliver, and Jason W. Chin. "A network of orthogonal ribosome· mRNA pairs." Nature chemical biology 1.3 (2005): 159-166.
- [18] Hartmann, K. U., and Charles Heidelberger. "Studies on fluorinated pyrimidines XIII. Inhibition of thymidylate synthetase." Journal of Biological Chemistry 236.11 (1961): 3006-3013.
- [19] Steidler, Lothar, et al. "Treatment of murine colitis by Lactococcus lactis secreting interleukin-10." Science 289.5483 (2000): 1352-1355.
- [20] Prakash, S., and T. M. S. Chang. "Microencapsulated genetically engineered live E. coli DH5 cells administered orally to maintain normal plasma urea level in uremic rats." Nature medicine 2.8 (1996): 883-887.