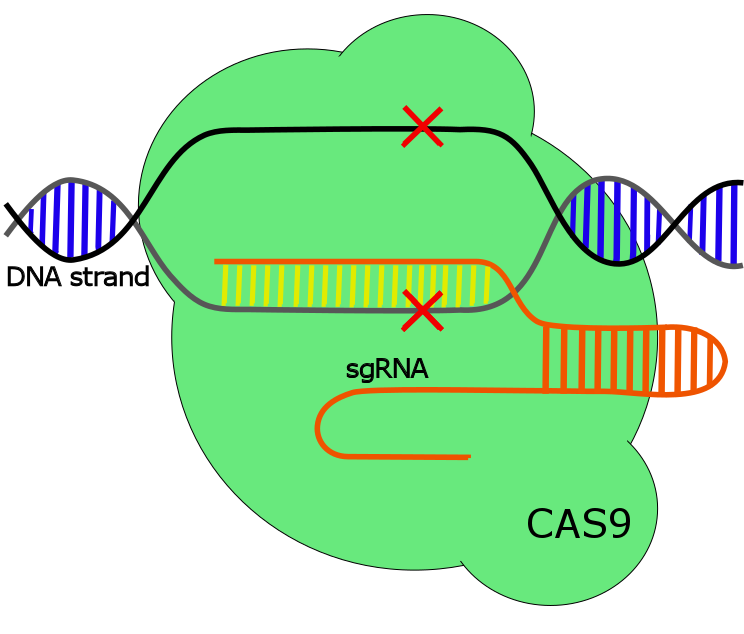
CRISPR/Cas9
The CRISPR system is an adaptive immune system response that some bacterial strains possess. Approximately 90% of microbes have a CRISPR design where the DNA sequence comprises of inverted repeats followed by approximately 30 random bases called spacer DNA. Several such stretches of DNA, each with different inverted repeats and spacer DNAs, are repeated which in turn gives rise to the name, Clustered Regularly Interspaced Short Palindromic Repeats, CRISPR.
The spacer DNA plays a vital role in recognizing bacteriophage DNA, here the microbe takes up phage DNA and preserves it for later use as a template for transcription of RNA guide molecules. In the type II bacterial CRISPR/Cas system the RNA molecules first gets cleaved into pre-crRNA. These RNA molecules later hybridizes with trans-activating crRNA, called tracrRNA, which possesses complementary sequences that match the crRNA:s short palindromic repeats.
When the formation of the RNA duplex occurs it in turn triggers RNase III, a dsRNA ribonuclease which processes and cleaves the RNA duplex to allow Cas9 binding. Cas9 is a nuclease with the objective to destroy target phage DNA. With the crRNA as the specific complementary marker to the target DNA sequence Cas9 accurately cleaving the DNA-helix on both strands. However, in the type II CRISPR system the target sequence will only be cleaved by Cas9 if it contains a Protospacer Adjacent Motif called the PAM sequence. PAM is a XGG sequence, where X can be any nucleotide. It acts to distinguish target DNA from CRISPR loci spacer DNA.
The described mechanism above is mainly a general description of the type II CRISPR system in its native form. However, to enable genome editing the crRNA-tracrRNA duplex in the type II system has been replaced with a fused synthetic version, called single guided RNA, sgRNA. sgRNA has demonstrated to be as effective and precise as the native system, but with the ability to specifically be designed to target specific genes (see figure 1) (1,2).
The Cas9 protein is purified from the bacteria Streptococcus pyogenes. The one used in our constructs is standardized and comes from iGEM. The guide RNA scaffold sequence used in this project is from IDT (3), and the target RNA sequence is from: Jiang W et al. 2014, Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii (4). This particular target RNA sequence have been used in C. reinhardtii before and targets the FKB12-gene causing rapamycin-resistance (4). This can consequently be used as a selection marker, indicating successful genome editing by Cas9/sgRNA (3).
The CRISPR/Cas9 technique open doors to new opportunities. A few examples are treatment and cure for diseases, development of hardier crops and modification of microorganisms for different purposes. The list of possibilities for CRISPR is long and only the imagination can set limits on areas of application.
References
1. A Modular Cloning Toolbox for the Generation of Chloroplast Transformation Vectors, Vafaee et. al (2014) https://www.ncbi.nlm.nih.gov/pubmed/25302695
2. Development and Applications of CRISPR-Cas9 for Genome Engineering, D. Hsu et. al (2014) http://zlab.mit.edu/assets/reprints/Hsu_PD_Cell_2014.pdf
3. Biolabs N. CRISPR/Cas9 and Targeted Genome Editing:A New Era in Molecular Biology | NEB [Internet]. Neb.com. 2016 [cited 23 September 2016]. Available from: https://www.neb.com/tools-and-resources/feature-articles/crispr-cas9-and-targeted-genome-editing-a-new-era-in-molecular-biology
4. Jiang W, Brueggeman A, Horken K, Plucinak T, Weeks D. Successful Transient Expression of Cas9 and Single Guide RNA Genes in Chlamydomonas reinhardtii. Eukaryotic Cell [Internet]. 2014 [cited 26 September 2016];13(11):1465-1469. Available from: https://www.ncbi.nlm.nih.gov/pubmed/25239977