
Mathematical Overview
With a knowledge of Michaelis-Menten enzyme kinetics, we were able to propose a model with appropriate parameters that are supported by data found in literature. Following on from previous MQ iGEM modelling, enzyme concentrations were derived for optimal production of chlorophyll a.

The initial velocity of the reaction (Vi) directly depends on the rate of the conversion of ES to P. It is also dependent on the total enzyme concentration [1]. By using the following formula, the total enzyme concentration and the speed of the reaction for the highest possible yield of chlorophyll a can be calculated from the values of Vi and S [1].
This is an example of 6 steps of the thirteen step pathway to chlorophyll a:

This reaction was translated into a mathematical equation which demonstrates the equations used for each enzymatic step used in the MATLAB models:

Our Models
The abbreviations used for the intermediates in all graphs are shown below. There are 13 intermediates/substrates for the 12 step pathway.
Abbreviations
- PPB - Porphobilinogen
- HMB - Hydroxymethylbilane
- URO - Uroporphyrinogen III synthase
- CPO - Coproporphyrinogen
- PPOX - Protoporphyrinogen IX
- PPIX - Protoporphyrin IX
- MgPPIX - Magnesium Protoporphyrin IX
- MgPPIXmono - Mg Protoporphyrin IX-mono-methyl ester
- Divi - Divinyl Protochlorophyllide
- Proto - Protochlorophyllide
- Clide - Chlorophyllide
- Chloro - Chlorophyll a
- ALA - Aminolevulinic acid
Chlorophyll a Pathway
The first precursor to this pathway begins with the tetrapyrrole precursor aminolevulinic acid (ALA), synthesised via the C-5 pathway. ALA is naturally produced in E. coli however we have also added it to increase the production of PPIX and hence more chlorophyll a. This precursor produces the colourless intermediate protoporphyrinogen IX (PPIX) where the tetrapyrrole biosynthetic pathway branches dependent on the binding of either iron to form heme involved in electron transfer for oxygen transport, or magnesium to form chlorophyll. E. coli naturally produce heme, this means they also produce ALA, however we will be supplementing this substrate to allow optimum yields of chlorophyll a. This pathway incorporates a magnesium cofactor with magnesium chelatase. The compound is then reduced by protochlorophyllide oxidoreductase to form a chlorin. The final step involves esterification of the alcohol phytol by chlorophyll synthase onto the remaining sidechain of chlorophyllide a, forming chlorophyll a. This year we have produced 3 models, the first reveals the amount of time required for the transformation of all ALA to the final product chlorophyll a.
The second model breaks down this pathway, showing the time required for the transformation of all ALA to form MgPPIXmonomethylester, the second last substrate in the pathway before the production of chlorophyll.
The third and final model produced from this years team followed the formation of MgPPIXmonomethylester from PPIX to see which enzymes are important for the speed of reaction.

Fig 1. Biosynthetic pathway from ALA to the first coloured intermediate protoporphyrin IX. a. ALA dehydratase; b. porphobilinogen deaminase; c. uroporphyrinogen III synthase; d. uroporphyrinogen decarboxylase; e. coproporphyrinogen oxidase; f. protoporphyrinogen oxidase [2].
Model 1
Chlorophyll a Theoretical Yield.
The yield of Chlorophyll a, and furthermore hydrogen, is dependent on the amount of PPIX supplied to the pathway, dependent on the concentration of ALA. ALA→ PPIX was previously modelled by the 2015 Mq iGem team. We have furthered their model to show the formation and depletion of substrates to produce chlorophyll a. We modelled this pathway using MATLAB to determine the highest yields of chlorophyll a we can achieve using E. coli as the expression host. Our model shows that if we input all necessary enzymes for the Chlorophyll a pathway at a concentration of 0.7 µM we get an ALA concentration of 1000µM, and after 16 days we have the maximum theoretical production of chlorophyll a (green line-see Figure. 2).

Fig 2. Model of the chlorophyll a pathway using MATLAB. Maximum chlorophyll a concentration of ~125 µM after 16 days with an input of 0.7 µM of enzymes and 1000 µM of ALA.
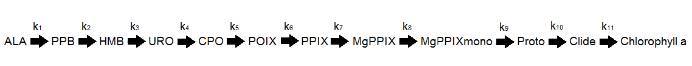
In this model we considered the following reaction for ALA to chlorophyll a transformation:
| Label | Enzyme | Function | Km (µM) | Kcat (1/s) | k1 | k-1 |
|---|---|---|---|---|---|---|
| K1 | Porphobilogen synthase | ALA → PPB | 1900 | 0.37 | 0.000197 | 0.0037 |
| K2 | Hydroxymethylbilane synthase | PPB → HMB | 7 | 0.105 | 0.0145 | 0.00105 |
| K3 | Uroporphyrinogin III synthase | HMB → URO | 5 | 500 | 101 | 5 |
| K4 | Uroporphyrinogin III decarboxylase | URO → CPO | 6 | 0.0039 | 0.000621 | 0.0000369 |
| K5 | Corproporphyrinogen oxidase | CPO → POIX | 2.6 | 0.00283 | 0.00110 | 0.0000283 |
| K6 | Protoporphyrinogen oxidase | POIX → PPIX | 7 | 0.292 | 0.0421 | 0.00292 |
| K7 | Magnesium chelatase subunit H | PPIX → MgPPIX | 3.2 | 0.00162 | 0.00005 | 0.0000162 |
| K8 | Magnesium protoporphyrin O-methyltransferase | MgPPIX → MgPPIXmono | 2.37 | 0.057 | 0.024 | 0.00057 |
| K9 | Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase | MgPPIXmono → Divi | 3 | 0.1 | 0.034 | 0.001 |
| K10 | Divinyl chlorophyllide a 8-vinyl-reductase | Divi → Proto | 3 | 0.1 | 0.00033 | 0.001 |
| K11 | Protochlorophyllide reductase and light-independent protochlorophyllide reductase subunit L | Proto → Clide | 1.84 | 0.1 | 0.055 | 0.001 |
| K12 | Chlorophyllide synthase | Clide → Chloro | 69 | 0.1 | 0.0015 | 0.001 |
Note: due to the lack of literature on these specific enzymes, the values for some of the later K values may be slightly different to theoretical values. For any future reproductions of the model or the experiment, we suggest running individual experiments of each K reaction for correct and accurate measurement of all values.
Model 2
ALA → MgPPIXmonomethylester.
Our second model estimated the time taken for ALA to be converted to MgPPIXmono. Input values of 0.7 µM of necessary enzymes, and a concentration of 1000 µM show that after a period of 8 days the PPIX intermediate levels begin to decline as it is converted to the next intermediate; MgPPIXmono (see Figure. 3-blue dotted line).

Fig 3. Model of ALA to MgPPIXmono using MATLAB, showing formation and depletion of intermediates. Input concentration of enzymes=0.7 µM, and ALA=1000 µM, at day 8 PPIX declines as it is forming MgPPIXmono to be further converted in the chlorophyll a pathway.
Model 3
PPIX → MgPPIX Yield.
From our first model of ALA to chlorophyll a, input values of 0.7 µM of necessary enzymes, over a period of 16 days 1000 µM of ALA was yielded (see Figure 2). However, the Chlorophyll a team conducted an experiment that fed 5000 µM of ALA to wildtype E. coli, expressed with MgCheletase plasmid. No detectable MgPPIX was produced, and only less than 5 µM amounts of PPIX were produced over 5 days. The low amounts of protoporphyrin fits the mathematical model.
It is noteworthy that the starting concentration of ALA fed to the pathway does not affect the production of chlorophyll a. Our third model of PPIX to MgPPIX shows that the maximum PPIX concentration after 8 days=100 µM, and that by adding more enzymes to this pathway, the rate of reaction is not increased.
There are 2 enzymes involved in this transformation of PPIX to MgPPIX. If enzyme 1 (magnesium cheletase) = 0.45 µM and enzyme 2 (magnesium protoporphyrin IX-methyltransferase) = 1 µM, the concentration of the PPIX substrate = 100 µM. Over the course of 8 days we get a concentration of 50 µM of MgPPIX, the consequent substrate in the chlorophyll a pathway (see Fig 4).

Fig 4. Model of PPIX to MgPPIX using MATLAB. Maximum production of 50 µM after 8 days with PPIX input of 100 µM. The 2 enzymes involved are enzyme 1 (magnesium cheletase = 0.45 µM) and enzyme 2 (magnesium protoporphyrin IX-methyltransferase) =1 µM.
References
- Keleti, T., & Kramer, M. (1986). Basic enzyme kinetics : Akademiai Kiado Budapest.
- Willows, R. (2004). Chlorophylls .





