Mito Morphin' Power Yeast Project Description
General Overview
Saccharomyces cerevisiae is used in a variety of valuable industrial processes. Its ability to grow and replicate utilizing aerobic respiration or fermentation, and its ability to survive without functional mitochondria, provide opportunities to control or alter mitochondrial function to optimize production processes or develop novel cellular subsystems in the mitochondrial space. To begin exploring these opportunities, we adapted two commonly used yeast expression vectors to the RFC 10 standard and used them to express yeGFP preceded by the mitochondrial localization signal from the mitoribosomal protein mRPS12. In addition, we attempted to demonstrate that aerobic respiration can be controlled by manipulating the expression of mRPS12. Without mRPS12, yeast mitochondria lack a functioning ribosome and become unable to produce proteins necessary for aerobic respiration. By expressing the mRPS12 protein in a haploid mRPS12 knockout strain, we investigated its usefulness as a means of regulating aerobic respiration in Saccharomyces cerevisiae.
Basic Trajectory and Parts
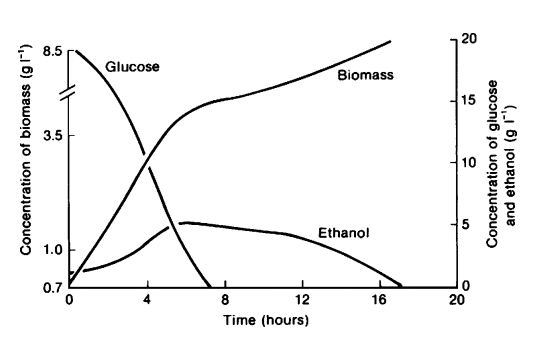
To compete with fast metabolizing prokaryotes, the surviving strategy of Saccharomyces cerevisiae is to quickly convert surrounding glucose into ethanol regardless of the presence of oxygen since yeasts are more tolerant to acid and ethanol [1]. Based on these characteristics, S. cerevisiae has become a widely used microorganism in food and beverage industries [2]. As clearly demonstrated by Figure 1 below [1], S. cerevisiae utilizes a “make-accumulate-consume” strategy regarding ethanol aerobically [3]. Once the glucose in the culture is depleted, ethanol production rate decreases due to continuous cell respiration. Therefore, typical alcohol fermentation processes are conducted in anaerobic conditions in order to prevent ethanol loss as indicated in Figure 1 [1]. To achieve a higher ethanol production rate, current industries often change yeast catabolism mechanism by altering external environment, for example adopting a fed-batch system. However, yeast grow much slower in anaerobic culture with less energy production [1]. Altering between fermentation and respiration by genetic engineering is potentially a better solution in this case. By gene manipulation, production process can start with aerobic respiration of yeast and switch to anaerobic fermentation when glucose level is low to prevent ethanol consumption. In this way, the amount of biomass as well as ethanol product will increase much quicker, and the peak ethanol concentration can be conserved.
Figure 1 Growth of S. cerevisiae on glucose in aerobic culture. When glucose becomes depleted at around 6 hours, yeast starts to consume ethanol as carbon sources [1].
One of the ways that aerobic respiration can be controlled in Saccharomyces cerevisiae is by manipulating the translation of electron transport proteins. Electron transport china proteins are translated by the mitochondrial ribosome, thus we propose that by controlling the expression of mitochondrial ribosomal proteins we can control aerobic respiration. Without the proteins encoded by mtDNA aerobic respiration stops [4]. We decided to investigate using expression of mitochondria ribosomal protein S12 in yeast as a means of controlling aerobic respiration. When the gene encoding mRPS12 is knocked out in BY4741 yeast, the strains exhibit a petite phenotype and become unable to utilize non fermentable carbon sources like glycerol [5]. We propose that without mRPS12 expression it is likely yeast will lose the ability to metabolize ethanol. Thus by manipulating the expression of mRPS12 we hope to optimize ethanol production in yeast.
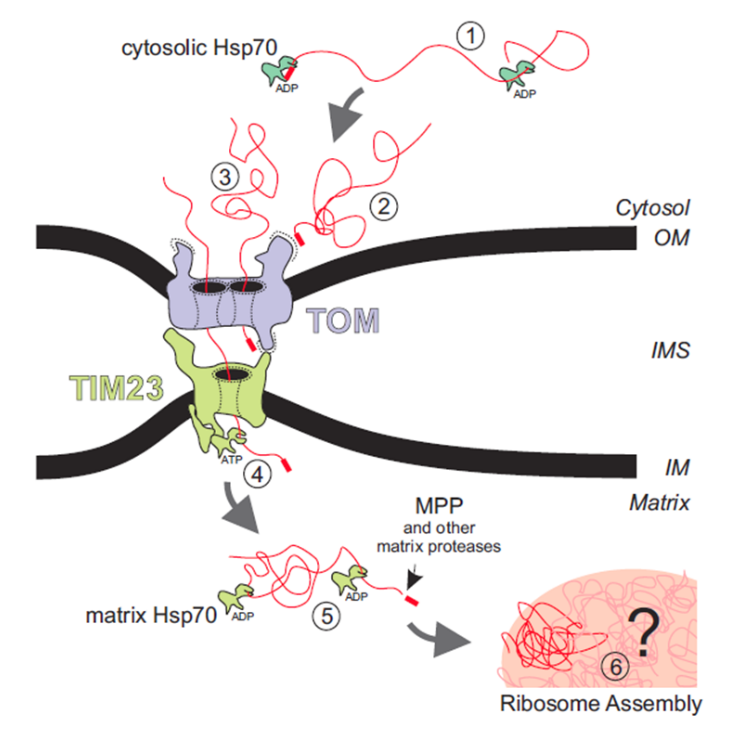
We also decided to verify the function of the matrix targeting signal portion of the mRPS12 gene. Mitochondria have very specific membrane proteins that will allow for proteins to be imported into the mitochondrial matrix. These two proteins are called the Translocase of the outer membrane of mitochondria (TOM) and the Translocase of the inner membrane of mitochondria (TIM). There are many different paths that proteins take, where it can be as simple as shuffling through both of the proteins. However, there are some other pathways where the protein goes through some processing in the intermembrane space.
Figure 2 Individual steps of the import reaction of protein into mitochondrial matrix [6].
As shown in Figure 2 above [6], Proteins that are imported to the mitochondrial matrix have a preprotein mitochondria localization signal or mls. This preprotein signal recruits chaperones and then binds to the receptor on the TOM complex. The protein is then translocated through the TOM pore, and then translocated through the TIM pore after which it is processed in the matrix. Commonly the preprotein signal is removed by MPP’s and other proteases [6].
We proposed that the function of the mRPS12 mls could be verified by creating a hybrid mls-yeGFP protein. yeGFP is a yeast codon optimized GFP protein, its excited by light in the wavelength of 395 to 485nm, and emits light at around 510 to 515 nanometers [7]. By joining yeGFP to the mls with a non-polar chain we hope to observe targeted areas of fluorescence inside yeast cells that might be the yeast’s mitochondria.
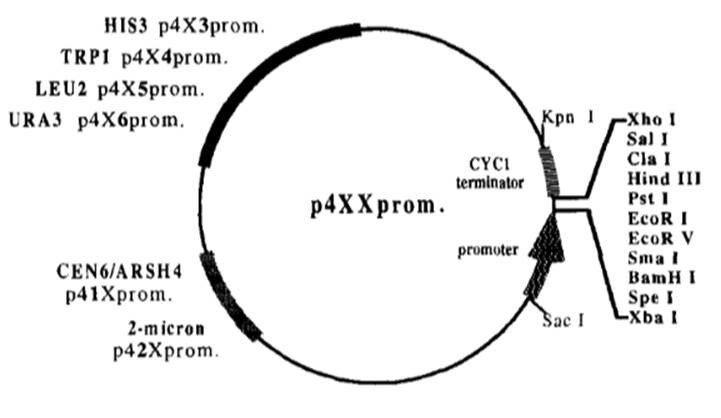
The two vectors we chose to convert were originally known as p413-GPD and p416-GPD both were produced as part of a 32 expression vector series [9]. The p4XX series was originally designed to allow comparisons between different levels of protein expression in different genetic backgrounds [9]. The series has vectors with different promoter strengths, different ORI’s that affect vector copy number, and different selectable markers but the same MCS [9]. We chose to convert p413-GPD and p416-GPD because they gave a middling level of gene expression, and their different selectable markers allow for plasmid shuffling.
Figure 3 Diagram of p4xx series plasmids showing key restriction sites and features [9].
References:
[1] J. Barnett and K. Entian, "A history of research on yeasts 9: regulation of sugar metabolism", Yeast, vol. 22, no. 11, pp. 835-894, 2005.
[2] M. Siddiqui, K. Thodey, I. Trenchard and C. Smolke, "Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools", FEMS Yeast Research, vol. 12, no. 2, pp. 144-170, 2012.
[3] E. Rozpędowska, L. Hellborg, O. Ishchuk, F. Orhan, S. Galafassi, A. Merico, M. Woolfit, C. Compagno and J. Piškur, "Parallel evolution of the make–accumulate–consume strategy in Saccharomyces and Dekkera yeasts", Nature Communications, vol. 2, p. 302, 2011.
[4] E. Nevoigt, "Progress in Metabolic Engineering of Saccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 72, no. 3, pp. 379-412, 2008.
[5] D. Sharma, A. Cukras, E. Rogers, D. Southworth and R. Green, "Mutational Analysis of S12 Protein and Implications for the Accuracy of Decoding by the Ribosome", Journal of Molecular Biology, vol. 374, no. 4, pp. 1065-1076, 2007.
[6] K. Fujita, T. Horie and K. Isono, "Cross-genomic analysis of the translational systems of various organisms", Journal of Industrial Microbiology and Biotechnology, vol. 27, no. 3, pp. 163-169, 2001.
[7] M. Woellhaf, K. Hansen, C. Garth and J. Herrmann, "Import of ribosomal proteins into yeast mitochondria 1", Biochemistry and Cell Biology, vol. 92, no. 6, pp. 489-498, 2014.
[8] B. Cormack, G. Bertram, M. Egerton, N. Gow, S. Falkow and A. Brown, "Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans", Microbiology, vol. 143, no. 2, pp. 303-311, 1997.
[9] D. Mumberg, R. Müller and M. Funk, "Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds", Gene, vol. 156, no. 1, pp. 119-122, 1995.