6/13/16
(HC) Made LB-Amp plates 450ml of DI water 5.0g tryptone 2.5g yeast 5.0g NaCl 7.5g Agar This was then adjusted to a pH of 7.0 using NaOH. It was then autoclaved. 2.5mL of 10mg/mL of Amp for a final concentration of 50 μg/mL was achieved.6/14/16
(HC) made CSM -His plates6/15/16
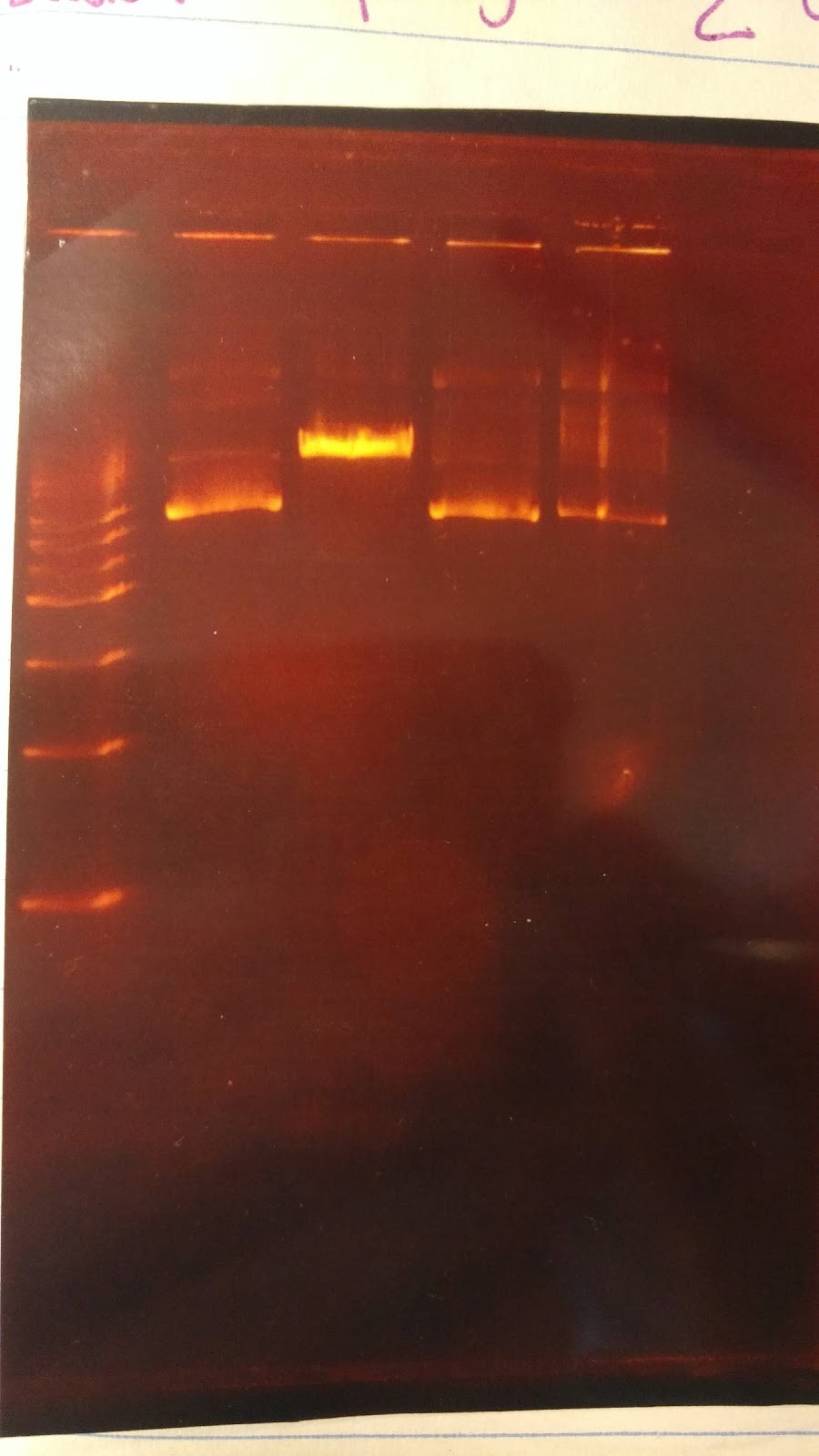
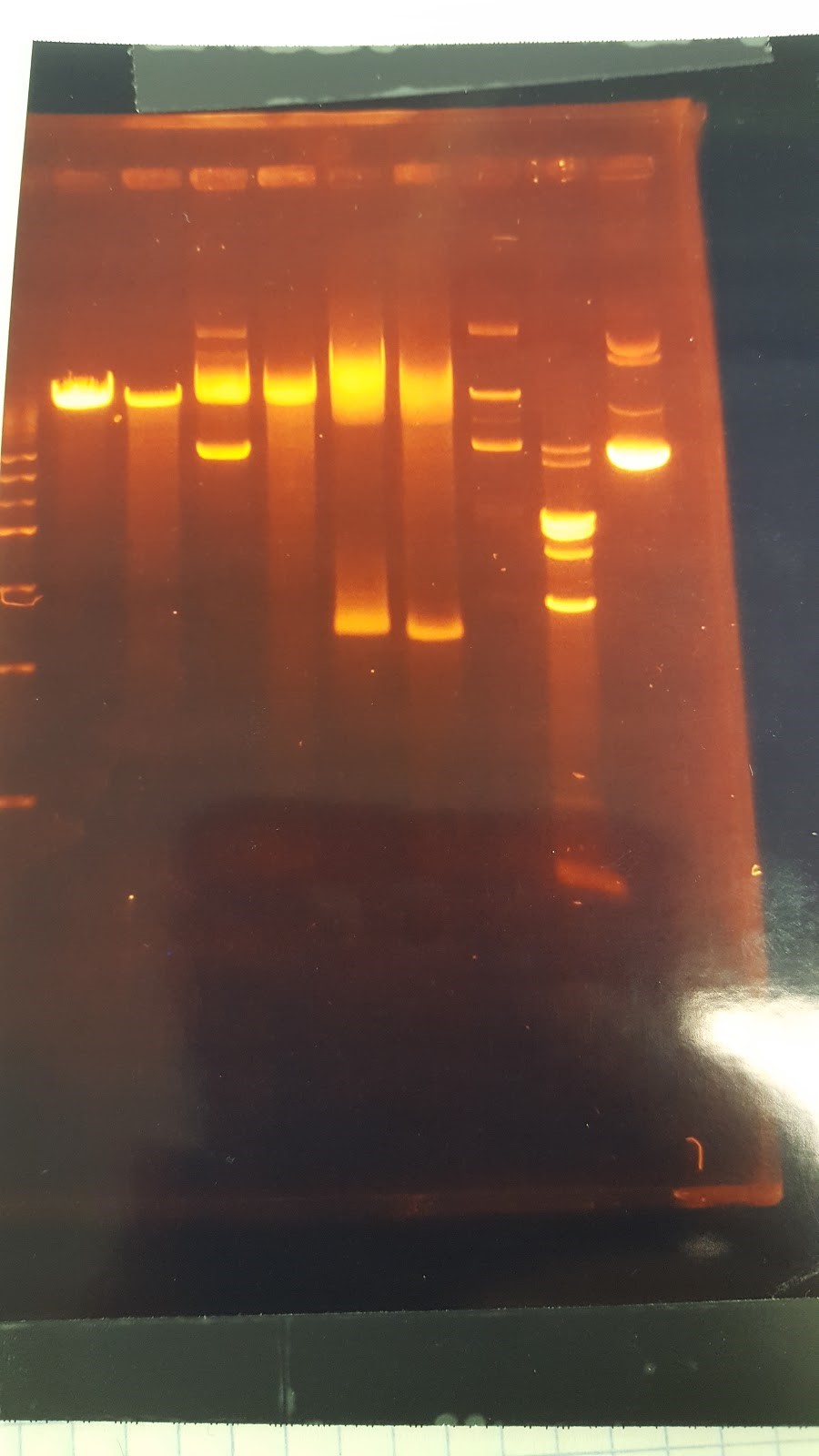
(XX) Goal: Run a gel to check whether pSB416 GPD has PstI site removed (diagnostic gel) 1. pSB416 GPD cut wit AvaI 2. pSB416 GPD cut with PstI 3. p146GPD cut with AvaI 4. p416GPD cut with PstI Major steps: Digest: 6 sterile water, 1 CutSmart buffer, 2 DNA, 1 enzyme (AvaI or PstI-HF) Gel Order: L, 1, 3, 2, 4Expected results on this diagnostic gel: 1. No cut because no AvaI site in pSB416 GPD 2. Should only see one cut because only one PstI site 3. Should see one cut 4. Should see two bands Experimental results: Gel results are not good; need to rerun this diagnostic gel Other Activities: Created mRPS12+/KanMZ- (parent) and mRPS12-/KanMX+ (mutant) master plate on YPD Replica plating YPD, YPG, -His, -Ura, -Ura+Glycerol, YPD+Paro, YPG+Paro
6/16/16
(HC) Ran a boiling and spin prep on P413 GPD Boiling Prep Procedure: 1) Pellet cells from 1.5mL of an overnight L-Amp culture of transformed E. coli cells in a microcentrifuge for 20-30 seconds and remove supernatant. 2) Use a toothpick and vortexer to resuspend the cells in 100 μl(0.1mL) of STET buffer(8% sucrose; 5%Triton X-100;50 mM EDTA; Tris-Cl, pH 8.0) containing 1mg/mL lysozyme(prepare 1 mg/mLenzyme in STET just prior to use; 1mL= 10 preps). 3) Place the suspension in a boiling water bath for 90 seconds. 4) Spin in a microcentrifuge for 15 min at the highest speed. 5) Remove the pellet (a gelatinous mass of cell debris) with a toothpick and discard. 6) Precipitate nucleic acid at -20℃ for 30 minutes using an equal volume of isopropanol. 7) Spin the tube in a microcentrifuge for 10 minutes at RTo to pellet the nucleic acids. 8) Was pellet twice with 70% ethanol and dry thoroughly. 9) Resuspend the pellet in 50 μL of sterile DI water or TE buffer. Spin Prep: We used the QIAprep Spin Miniprep Kit to run a spin prep and followed the instructions inside. (XX)Goal: Rerun the diagnostic gel stated yesterday (6/15/16) Experimental results: Good gel results, confirming that the PstI site has been removed in pSB416 GPD. pSB416 ready to be sequenced. Other activities: Incubated pSBIC3 mRPS12 TU and pSBIC3 mRPS12 mls in LB-chloramphenicol broth @37C overnight at 250 rpm6/17/16
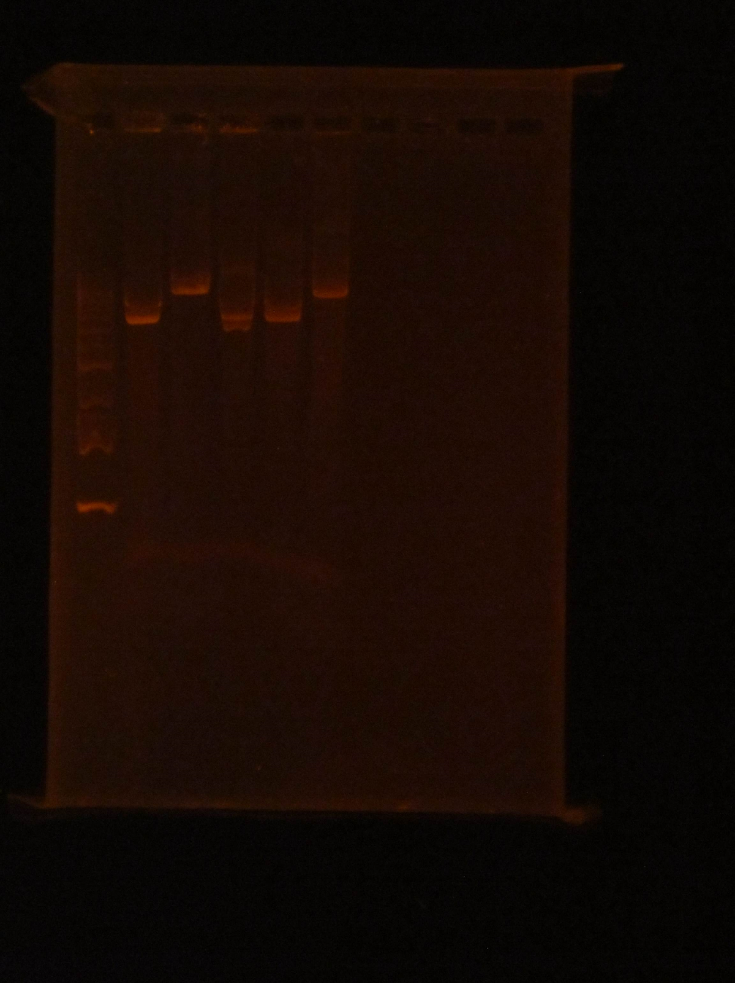
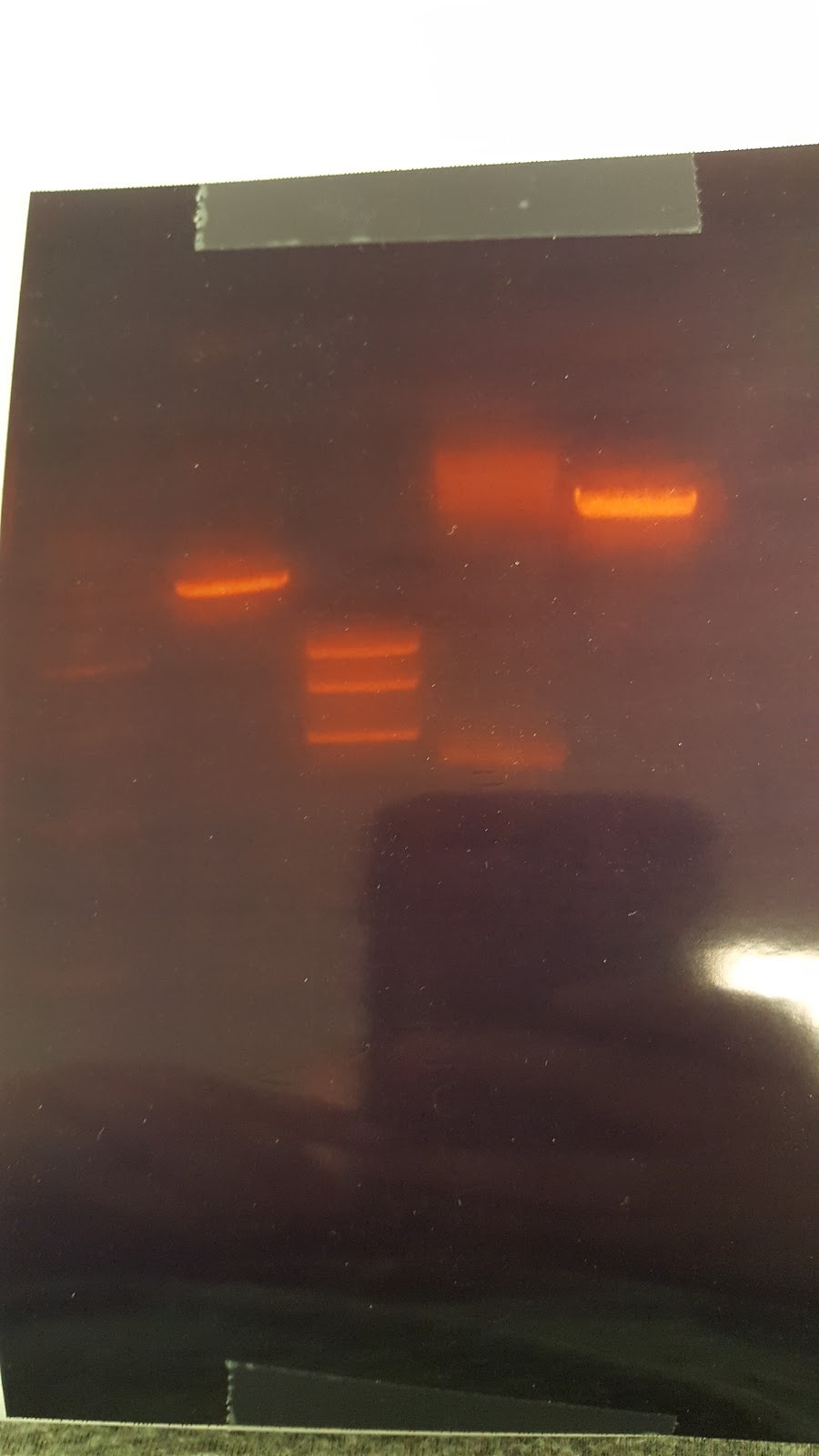
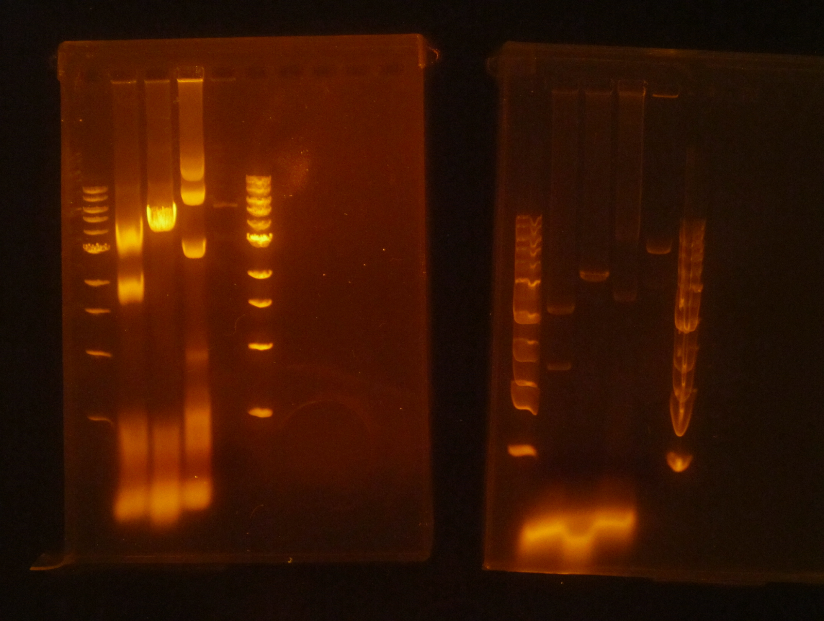
(HC) Ran two digests for P413 GPD Analytical Digest: 5 μL of DNA(Boiling Prep) 1μL of PstI HF 1μL of CutSmart buffer 3μL of sterile DI water Preparative Digest: 7μL of DNA(Spin Prep) 1μL of NheI 1μL of NsiI 1μL of CutSmart buffer Gel Results: PREP GEL Lane 1: Ladder Lane 3: Preparative DigestLane 1: Ladder Lane 2: Uncut (2μL of DNA and 8μL of water) Lane 3: Analytical Digest
(XX) Master plate results: Nothing grew on control plates (-His, -Ura, -Ura+Glycerol) Both parent and mutant grew on YPD and YPD+Paro, indicating that Paro has no effect on yeast growth Mutant cannot grow on YPG or YPG+Paro because of the mRPS12 gene knockout Other Activities: Cut p413GPD with NheI and NsiI to get rid of the illegal PstI site Massive gel to run boiling preps Conclusion: P413GPD cut looks like an uncut plasmid Massive gel has many blurry bands, which indicates that boiling preps are very impure
6/20/16
(XX) Ultimate Goal: Put pSBIC3 mRPS12 TU into our own standardized yeast vectors Validate mls function by creating mls-yeGFP construct Goal for today: Digest and Harvest mRPS12 TU from pSBIC3(tube 1) 5μL of DNA 1μL of EcoRI-HF 1μL of pstI-HF 1μL of CutSmart buffer 2μL of sterile water Digest and Harvest mRPS12 mls from pSBIC3 (tube 2) 5μL of DNA 1μL of EarI 1μL of SpeI 1μL of CutSmart buffer 2μL of sterile water Gel Order: L, 1C, 1XX, 1XL, 2C, 2XX, 2XL **C-uncut plasmid, XX-Xintong, XL-Xander Gel results: Uncut does not show up on the gel Not a very good results but we harvested the bands anyway Other activites: Excise bands from the gel and perform gel extraction (QIA kit): Tube 1 = 0.26 g Tube 2 = 0.14 g Tube 3 = 0.17 g *tube 3 contains p413 GPD with NsII/NheI cut Prepare Yeast Competent Cells 10 mL YPD, inoculated at 250RPM @ 30C Both BY4741 and BY4741 mRPS12 is prepared6/21/16
(XX) Goal for Today: DNA Assembly Major Steps: We used nanodrop to determine the amount of DNA in tube 1 (TU) and tube 2 (mls) Tube 1: 4.5 ng/μL Tube 2: 4.0 ng/μL * Nano drop results indicate that we need to precipitate DNA for further experiment After precipitation, nanodrop reports 12.6 ng/μL DNA NEBuilder HiFi DNA Assembly Cloning Kit: 2μL Ear/SpeI digest pSBIC3 mls 8μL fragment 16-4 (yeGFP) 10μL HiFi Master *All three above, set the reaction on ice; 1:2 vecotr: insert Thermocycler Other Activities: Yeast competent cell from yesterday did not grow Prepare new yeast competent cells: 5 mL YPD in round bottom tubes6/22/16
(XX)Goal: Prepare Yeast Competent cells Diagnostic gel on mRPS12 TU minipreps Major Steps: Yeast competent cells: Spectromphotometer OD 260: 1000 YPD Blank 1000 KnockOut(KO) has 1.777 Abs. 1000 Parent has 2.222 Abs. Regrow these cells into 15 mL broth for about 2 hrs: Take 0.5 mL current culture into 14.5 mL YPD Spectrophotometer Results: KO: 0.390 Abs. Parent: 0.692 Abs. Follow yeast competent cell protocol Diagnostic Gel: 1: pSBIC3 mRPS12 TU miniprep 1 cut with EcoRI and PstI 2: pSBIC3 mRPS12 TU miniprep 2 cut with EcoRI and PstI 3: pSBIC3 mRPS12 TU miniprep 1 cut with PstI 4: pSBIC3 mRPS12 TU miniprep 2 cut with PstI Gel Order: L, 1, 2, 3, 4 Gel Results: For #2, fragment should be around 500bp Gel results indicate that we should redo minipreps or bad enzymes6/23/16
(XX) Goal: Test if the enzymes are still okay to work with Redo miniprep at the same time Major steps: Enzyme test: Tube 1: EcoRI-HF Tube 2: PstI-HF Tube 3: SpeI-HF Tube 4: EarI Gel Order: 1, 2, 3, 4, L Gel Results: enzymes are working properly6/24/16
(HC) After extracting P413 GPD fragment from the gel, I precipitated the DNA Precipitating DNA: 1.)Add 1-2 μL of 5M NaCl to 30 μL of DNA 2.)Add 62 μL of ice cold EtOH 3.)Let sit on ice for 30 minutes 4.)Centrifuge @ 0℃ for 10 minutes at 13 krpm 5.)Remove supernatent 6.)Fill tube to halfway point with 70% EtOH 7.)Centrifuge @ 4℃ for 2 minutes 8.)Remove supernant 9.)Let dry 10.)Add 6μL of Sterile Water. Obtained concentration of DNA with NanoDropper
| Nucleic Acid Concentratione | A @ 260 nm | A @ 280 nm | 260/280 | 260/230 |
|---|---|---|---|---|
| 75.7 ng/μL | 1.514 | 0.922 | 1.64 | .033 |
| 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| 416 control | 426 control | 416 CYC1 | 416 CYC1 | 426 GPD | 426 GPD |
The gel shows that the results we not what we wanted due to there being two bands instead of 1 band showing the Pst1 site was not removed. (XX) Talked about light switch design LexA promoter instread of Gal promoter PhyB/PIF3 red light system
6/25/16
(XX) Miniprep mRPS12 TU and mRSP12 mls(total of 8 minipreps)6/27/16
(XX) Goal: Redo digest and harvest TU and mls Major steps: Cut mRPS12 TU with EcoRI/PstI Cut Mrps12 mls with EarI/SpeI Gel order: L,TU1a, TU1b, TU2a, TU2b, mls1a, mls1b, mls2a, mls2b Gel results: harvested TU bands from TU2a and TU2b (2 bands per tube) TU gel extract = 0.363 g harvested four mls bands (2 bands per tube) mls gel extract A = 0.342 g mls gel extract B = 0.255 g Other activities: QIA gel extraction kit Used nanodrop to test DNA amount: TU = 3.1 ng/μL mls A = 41.2 ng/μL mls B = 27.7 ng/μL *according to nanodrop readings, TU needs to be precipitate while we run NEBuilder on mls A and mls B (HC) Ran NEBuilder on P413 GPD and 16-1 fragment. 1μL DNA 3μL fragment 10μL NEBuilder HiFi DNA Assembly Master Mix 6μL Sterile DI water (JM/CH) The first thing we are doing is running the blunt end ligation protocol using NEB Quick Blunting Kit and the NEB Quick Ligation Kit This was done with 3 tubes of 416 CYC1 and 3 tubes of 426 GPD Next we did a transformation on one of the 416 CYC1 and one of the 426 GPD Those are spread on plates and are now incubating so we can use them tomorrow6/28/16
(JM/CH) Slow day today because all we can do is inoculate falcon tubes for 24 hours so we can run a mini-prep tomorrow We did 6 tubes of 416 CYC1 And 6 tubes of 426 GPD (XX) Goal: NEBuilder TU perform transformation for TU, mls A, and mls B Redo TU digest to check TU1a, TU2a: just PstI enzyme TU1b, TU2b: double digest (PstI/EcoRI) Redo NEBuilder mls A and mls B *due to programing issue?? Gel order: L, Uncut (TU), PstI TU1a, PstI/EcoRI TU1b, PstI TU2a, PstI/EcoRI TU2b Gel results: the band after only PstI cut seems too long – questioning the pSBIC3 mRPS12 TU stock??6/29/16
(JM/CH) After incubating for an undetermined amount of time due to our shaking incubators being complete pieces of shit we are now going to mini preps because we could not find the proper buffer for the boiling preps With the miniprep material we had we were able to do 3 tubes of 416 and 2 tubes of 426 Also made more LB Amp Ran the digest of the plasmids with Pst1-HF (XX) Transformation results: No transformants for mls Major steps: Check mls minipreps Cut with only EarI and run gel Other activities: Streak pRS416 (the vector with no promoter) on LB-Amp plates – for LexA light switch Transform competent E. Coli with pSBGPD416 A, plate on LB-Amp Gel order: L, mls A uncut, mls 1a cut Gel results: bad mls miniprep – questioning the mls stock??6/30/16
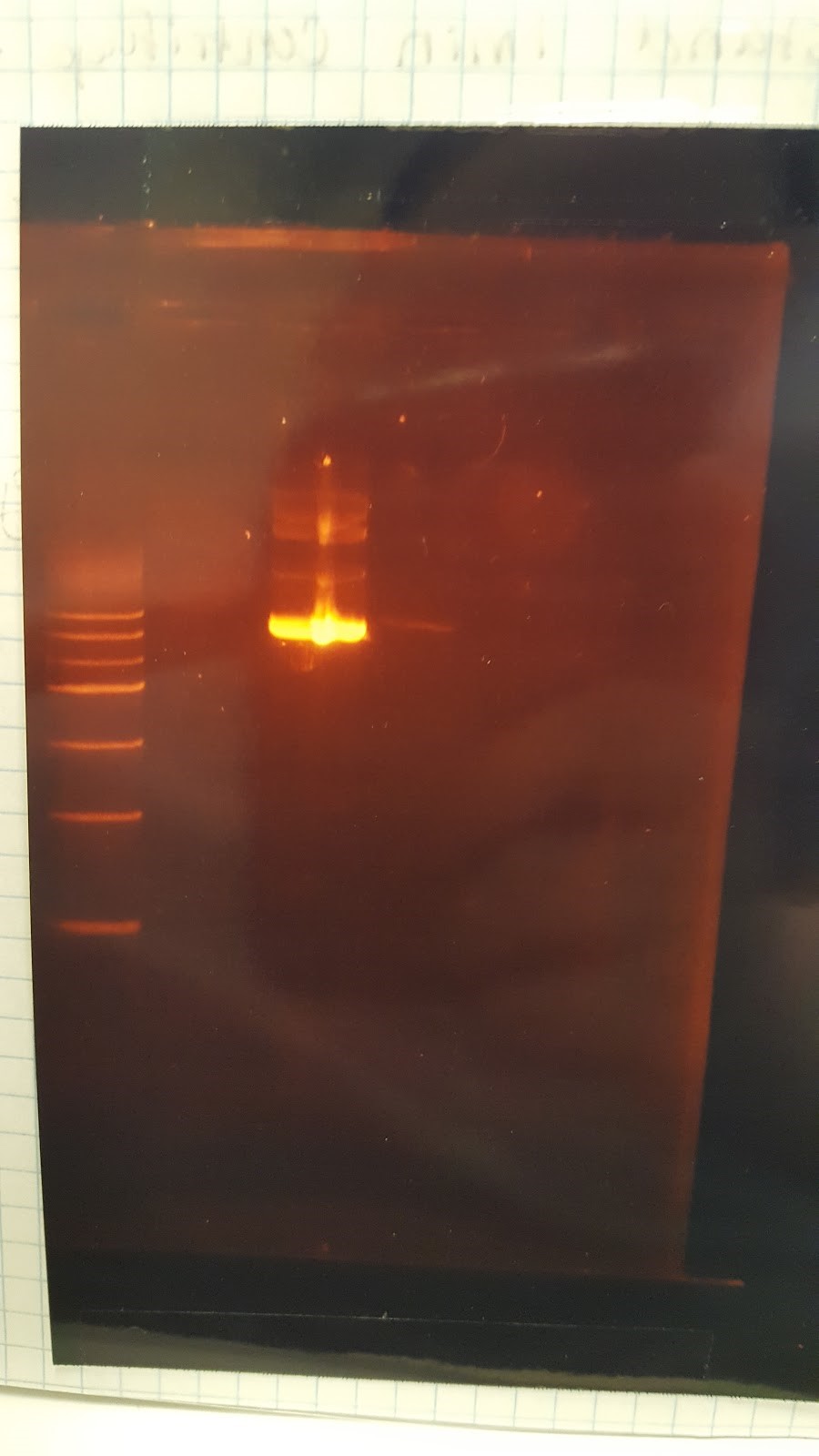
(JM/CH) Today we are running the gel of the digest that should result in seeing only one Pst1 site Gel Set-up Ladder / 416 1 / 416 2 / 416 3 / 426 1 / 426 2 Results: too blurry to come to any reasonable conclusionNow to set up a proper gel to run tomorrow We need to set up two controls, which will be minipreps of the DNA plasmids with 2 Pst1 sites Also we will be using 5 of our blunt end ligated DNA (3 416, and 2 426) We will then also load two of the wells, one with 416 and one with 426, but uncut (XX) Found out that pSB416GPD I from last year has failed because it has an extra start codon before BB prefix Goal: extract mRPS12 TU from old pSB416 GPD I Major steps: Made a 20 digest – 10 to run gel and 10 to run NEBuilder 10 pSB416 GPD I from last year 1 EcoRI 1 PstI 2 CutSmart Buffer 7 sterile water Other activities: Streak pSBIC3mls I on LB-chloro Inoculate pSB416 GPD A in LB-Amp Inoculate pRS416 in LB-Amp Gel Results: Nice mRPS12 TU band on the gel, extract mRPS12 TU band Mass = 0.265 g Nanodrop results TU: 5.6 ng/μL *need to do precipitation
7/1/16
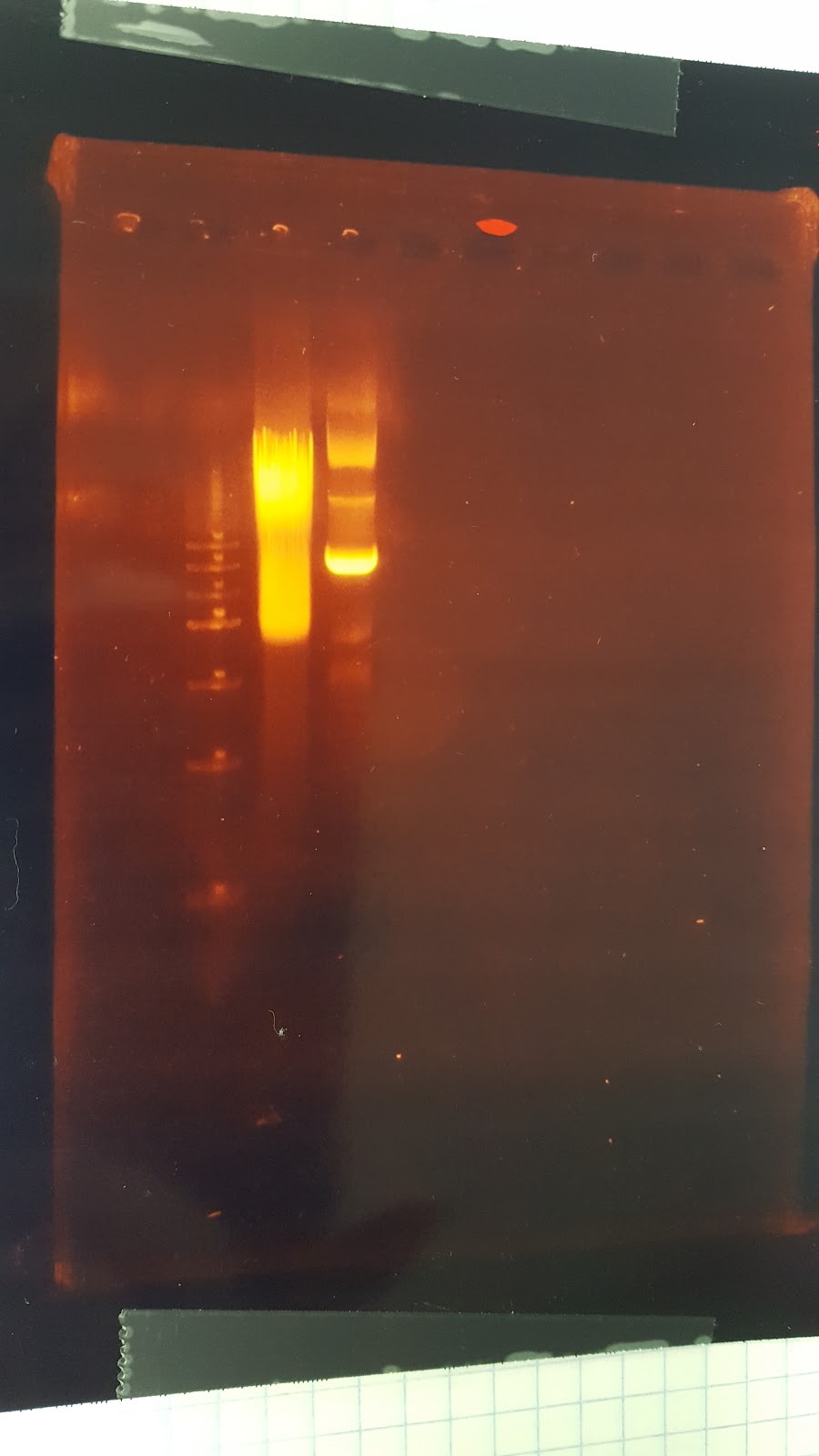
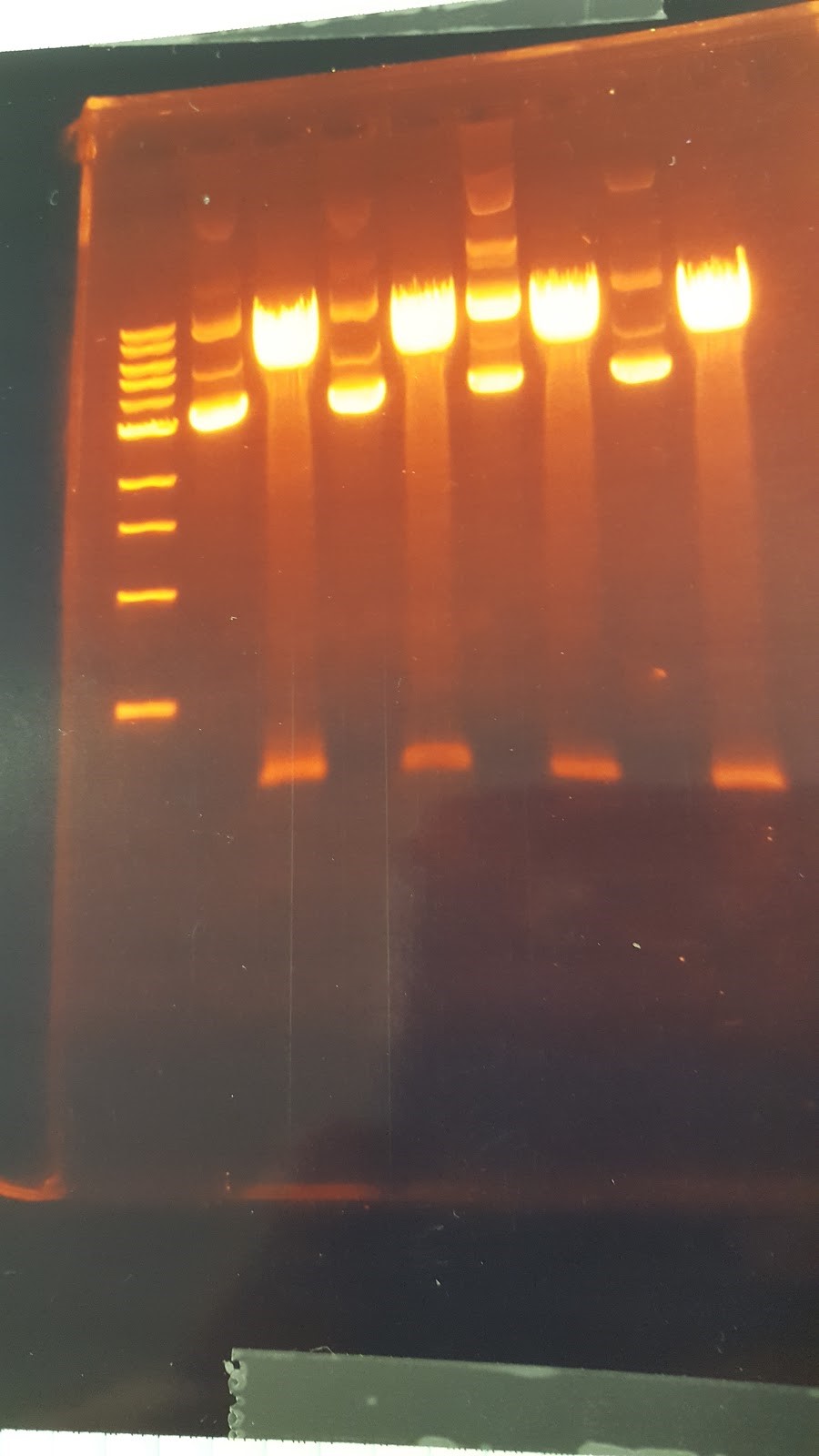
(HC) Ran a spin miniprep using the Monarch Nucleic Acid Purification Kit and the instructions inside on the P413 Transformant. Ran a digest 7μL of DNA 1μL of Cutsmart 1μL of PstI 1μL of Sterile water Ran gel with cut and uncut transformant Lane1: Ladder Lane 2: Cut Lane 3: Uncut(JM/CH) Today we ran the gel that had out plasmids hopefully set for perfection ladder / 416 1 / 416 2 / 416 3 / 416 orig / 416 uncut / 426 uncut / 426 orig / 426 1 / 426 2 Results:
This shows that our gel yesterday is confirmed and every sample shows one band. The Pst1 worked due to the double bands in wells 5 and 8. Edit: After talk with PI the gel does not have correct size of bands. Still inconclusive. (XX) Precipitation of TU from yesterday resulted 3.8 ng/μL according to nanodrop. This result does not make sense and it might possibly due to nanodrop defecting DNA after gel isolation. So we continue on NEBuilder anyway. Other activities: miniprep pRS416 and pSB416 GPD A inoculate pSBIC3 mls in LB-chloro broth
7/2/16
(XX) Goal: Miniprep pSBIC3 mls TU transformation7/3/16
(XX)We see transformants on 7/3/16! This is exciting news!7/5/16
(HC) Ran a digest with P413 GPD 2 μL of DNA 1μL of Cutsmart 1μL of PstI 6μL of Sterile Water Ran Gel *** Ran a couple different experiments on one gel and will only list the ones related to P413 GPD experiment, the others will be listed in another Lab members notebook section Lane 1: Ladder Lane 2: Cut Lane 3: UncutRan Digest on P413 GPD on two different preps *Prep was run by Connor #Prep was run by Holly Both Preps were run through the same digests 2 μL of DNA 1 μL of NheI 1 μL of 2.1 buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of NsiI 1 μL of Cutsmart buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of PstI 1 μL of Cutsmart buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of EARI 1 μL of cutsmart buffer 6 μL of Sterile Water (XX) Goal: Diagnostic gel of pSB416 GPD mRPS12 TU: used HindIII or PstI enzymes Isolate mls DNA from mRPS12 mls miniprep: double digest using EarI and SpeI Gel Order: L, pSBIC3 Cut, pSBIC3 Uncut, B, mls digest, mls uncut, B, TU PstI, TU HindIII, TU uncut Gel Results: TU with HindIII has weird result, possibly due to star activity or failed digest? Redo pSBIC3 Redo mls Redo TU digest Other activities: Made master plate on CSM-D-URA pSB416 GPD A mRPS12 (5-8) pSB416 GPD A Parent (1-4)
7/6/16
(HC) Ran Gel Ladder / NheI* / NheI# / NsiI* / NsiI# / PstI* / PstI# / Ear1* / Ear1# / UncutRan digest on P413 GPD of Holly’s prep(# prep from above) 2 μL of DNA 1 μL of EarI 1 μL of Cutsmart buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of XhoI 1 μL of Cutsmart buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of PstI 1 μL of Cutsmart buffer 6 μL of Sterile Water Ran gel of these digests Ladder / Uncut / Ear1 / PstI / XhoI
7/7/16
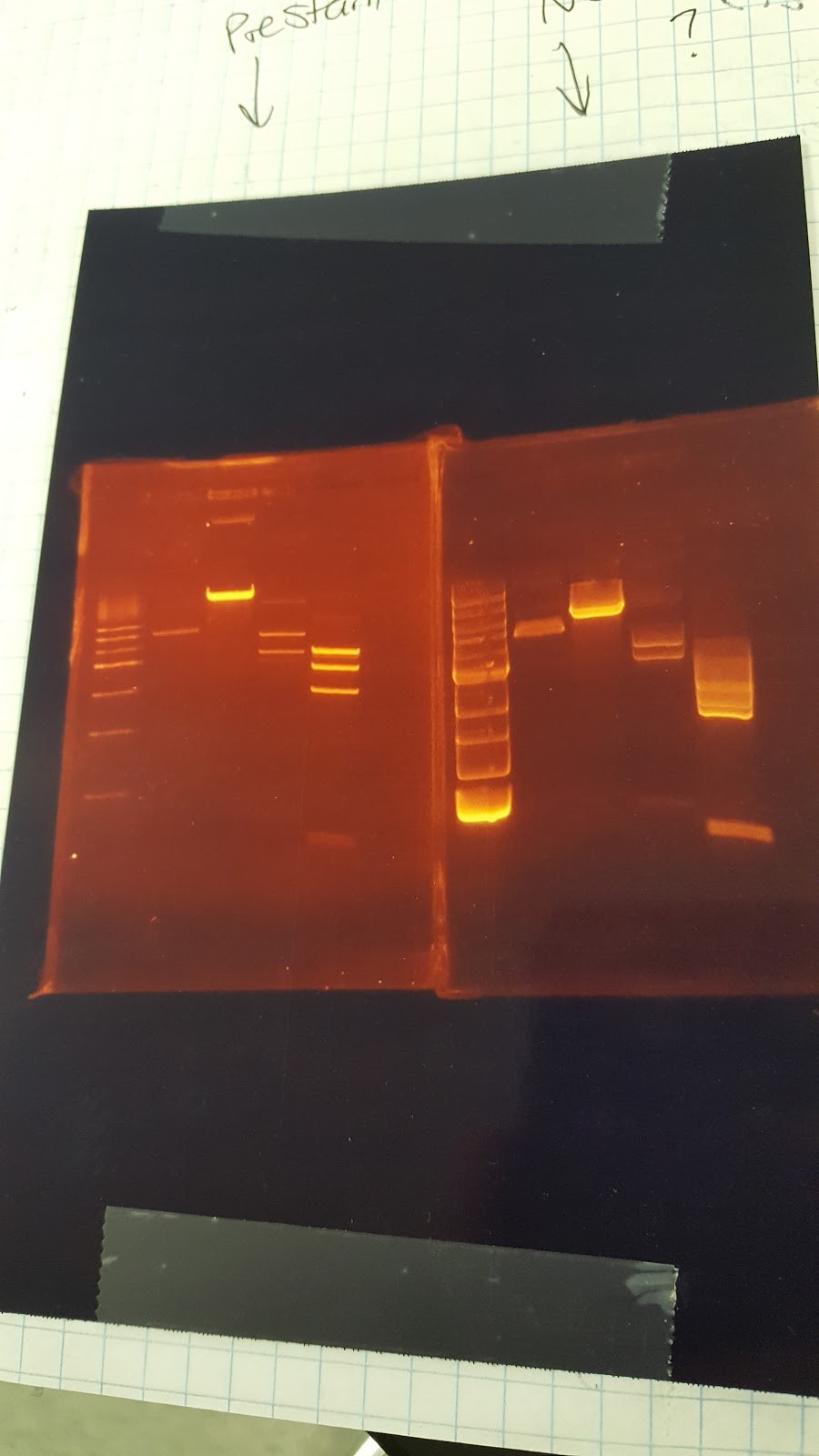
(HC) Ran digest w/ EarI and XhoI on my preps for P413 GPD(*) and a team member’s prep of P413 GPD(#) from last year. 2 μL of DNA 1 μL of EarI 1 μL of Cutsmart buffer 6 μL of Sterile Water 2 μL of DNA 1 μL of XhoI 1 μL of Cutsmart buffer 6 μL of Sterile Water Ran a gel to compare the preps of P413 GPD and the effects of Pre-staining vs. Post-Staining. Gel 1: Pre-stained 500bp Ladder / XhoI # / XhoI* / EarI# / EarI* Gel 2: Not stained (we hadn’t stained this gel yet but it was clear to see bands) 1kbp Ladder / XhoI # / XhoI* / EarI# / EarI*(JM/CH) Inoculated tubes that we will use tomorrow with 416 CYC1 and 426 GPD Made up 50mL STET buffer • 4g 8% Sucrose • 2.5 mL 5% Trition X-100 • 5 mL 50 mM EDTA • 2.5 mL 50 mM Tris-Cl
7/8/16
(JM) Alpha and beta are different designations to distinguish between two possible candidates for testing. FIRST ROW: ladder / 416 pst1 alpha / 416 EcoR1 alpha / 416 Uncut alpha / 416 Uncut beta / 416 Pst1 beta / 416 EcoR1 beta SECOND ROW : ladder / 426 pst1 alpha / 426 EcoR1 alpha / 426 Uncut alpha / 426 Uncut beta / 426 Pst1 beta / 426 ecoR1 betaChose Both 416CYC1 alpha and 426 GPD alpha to freeze away and make new cryo tubes (XX)Goal: Redo pSB416 GPD mRPS12 TU diagnostic gel Double digest pSBIC3 mRPS12 mls Gel Order: L, TU, B, mls uncut, mls cut Gel results: TU has no insert, need to restart from NEBuilder Can isolate mls cut band and do gel extraction Other activities: NEBuilder to form mls-yeGFP construct: 1 Digest 9 16-4 fragment **Check for time-saver qualified enzymes – only need to digest for 5 to 15 minutes
7/17/16
(HC) Made Amp plates and CSM-URA plates7/18/16
(HC) Plated P413 GPD onto AMP-Plates to allow to grow for colonies. (JM/CH) Today we are going to run two gels to test if prestaining or post staining is more effective The gel inside the clear container while the nonstained one is in the blue one Ladder / 416 CYC1 cut w/ Pst1 / 416 CYC1 cut w/ EcoR1 / 416 CYC1 Uncut / mRPS12 tu invalid / Ladder #2The gel on the right was not stained in any manner, but bands can still be easily seen. (XX) Goal: Transformation for pSBIC3 mls-yeGFP on LB-Amp plate
7/19/16
(HC) Inoculated LB Amp tubes with colonies from platesI selected two colonies from each plate and labeled them 1A, 1B, 2A, and 2B based on which plate and colony it was. (JM/CH) Today the plates have grown up nicely. We picked 3 colonies from each of the 2 plates (one 416 and one 426). Now they are growing up in the incubator. Creating lesson plan. (XX) Transformation from yesterday did not work because of the LB-Amp plates! **pSBIC3 is chloro selective!! Goal: Plate NEBuilder stuff from 7/8/16 on LB-chloro plates
7/20/16
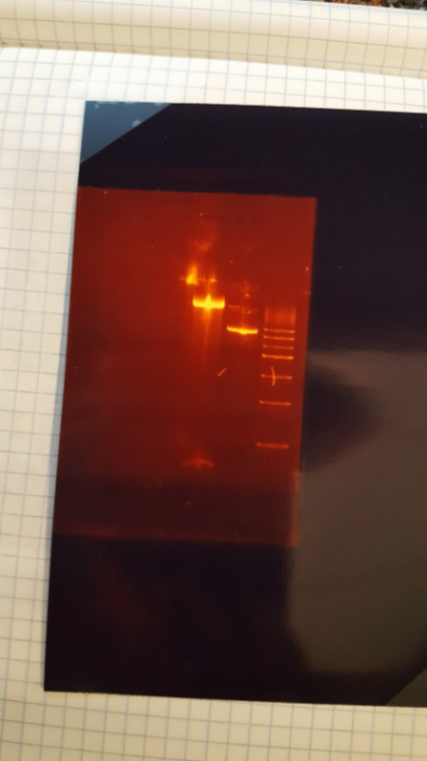
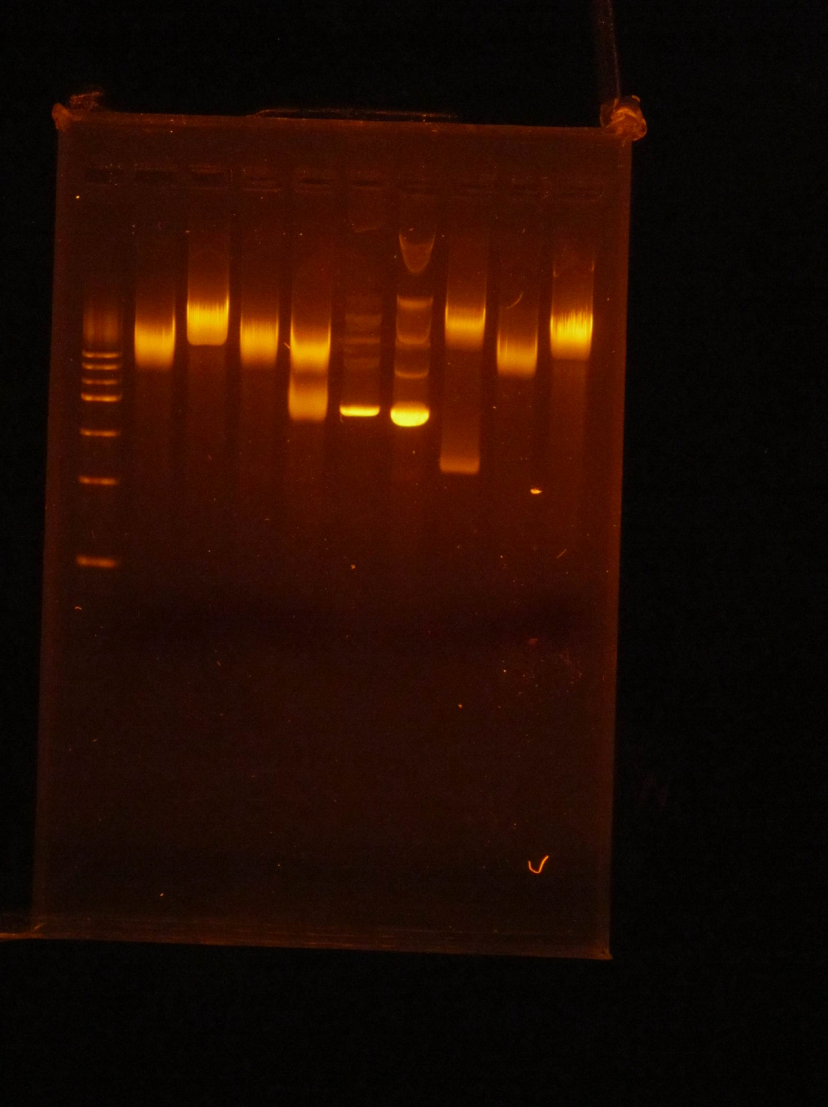
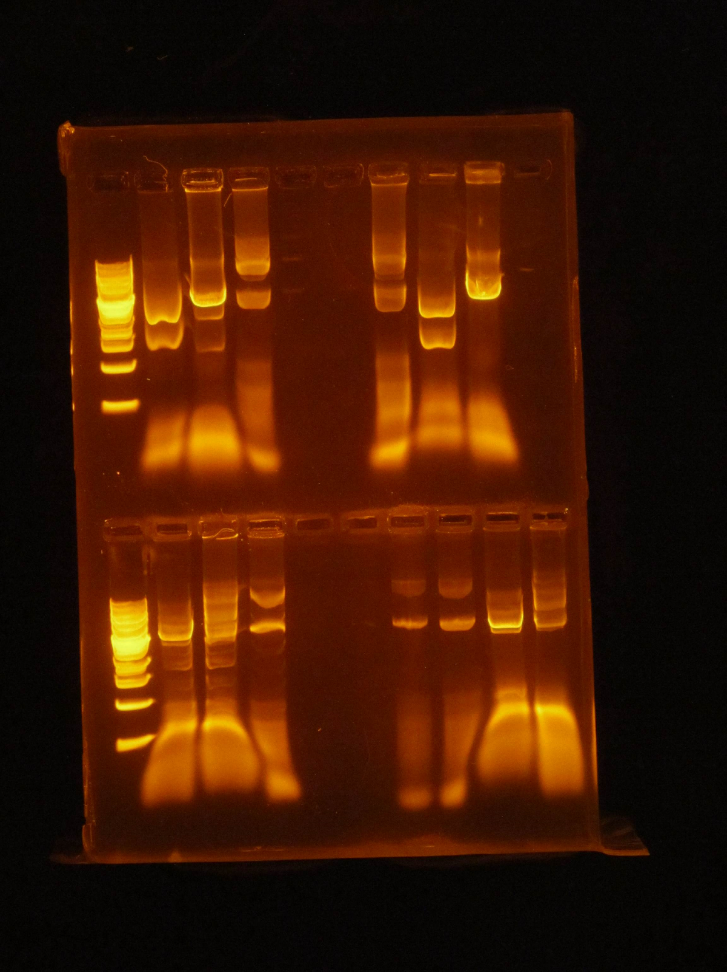
(HC)Ran a miniprep on all 4 tubes that were inoculated with the cells from the P413 GPD plates, 3 minipreps for each tube for a total of 12 tubes. I used the Monarch Plasmid Miniprep Kit. Digested 1A, 1B, 2A, and 2B with NheI and NsiI 5 μL of DNA 1 μL of NheI 1 μL of NsiI 1 μL of Cutsmart buffer 2 μL of Sterile Water I ran a gel with these digests. 1kbp Ladder / 1A uncut / 1A cut / 1B uncut / 1B cut / 2A uncut / 2A cut / 2B uncut / 2B CutI cut out the bottom bands of Lanes 3, 5, 7, and 9 and kept them to eventually do a gel extraction. (JM/CH) Today we started off the day by doing mini preps to isolate our plasmids. Helped Holly with her 413 GPD miniprep. Worked on the 416CYC1 and 426 GPD miniprep Now we are going to do the blunt end ligation to cut out the extra Pst1 site in both the 416CYC1 and 426 GPD plasmids. The gel came back from the miniprep of 413 and can be seen above in Holly’s entry. Now we are finishing the blunt end ligation of the 416 and 426. We kept tubes of just the blunted DNA and we will have a stock of the ligated DNA as well. (XX) There are still no transformants for mls-yeGFP! L Goal: Miniprep TU transformants and diagnostic gel to check if we have any insert Redo mRPS12 mls double digest – start mls-yeGFP process from gel extraction Gel mass = 0.286 g Gel Order: L, A1, A2, A3, A4, B1, B2, Blank, mls uncut, mls cut (A1 to B2) are minipreps from TU transformants Gel Results: Gel shows TU transformants have no insert
7/21/16
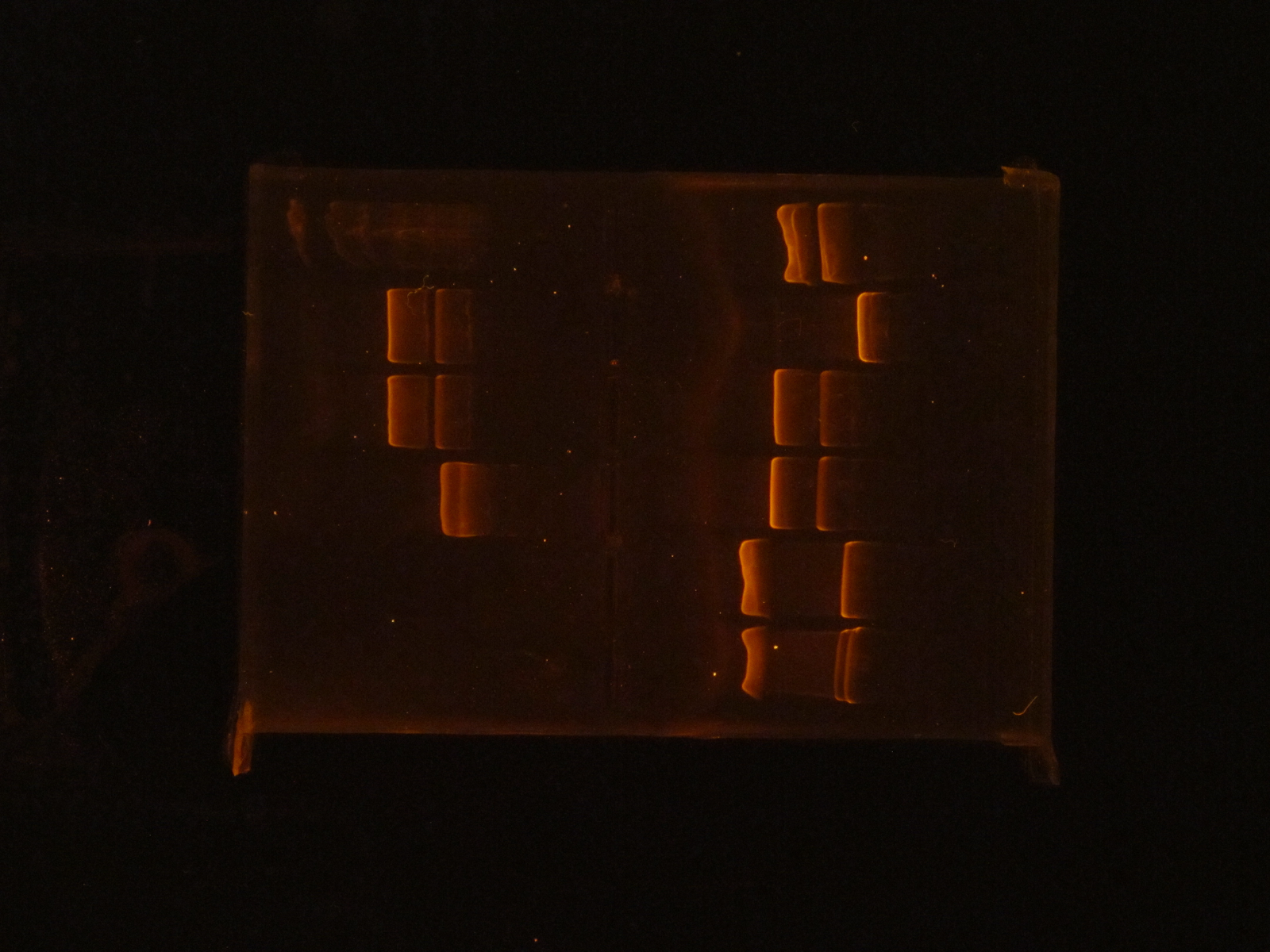
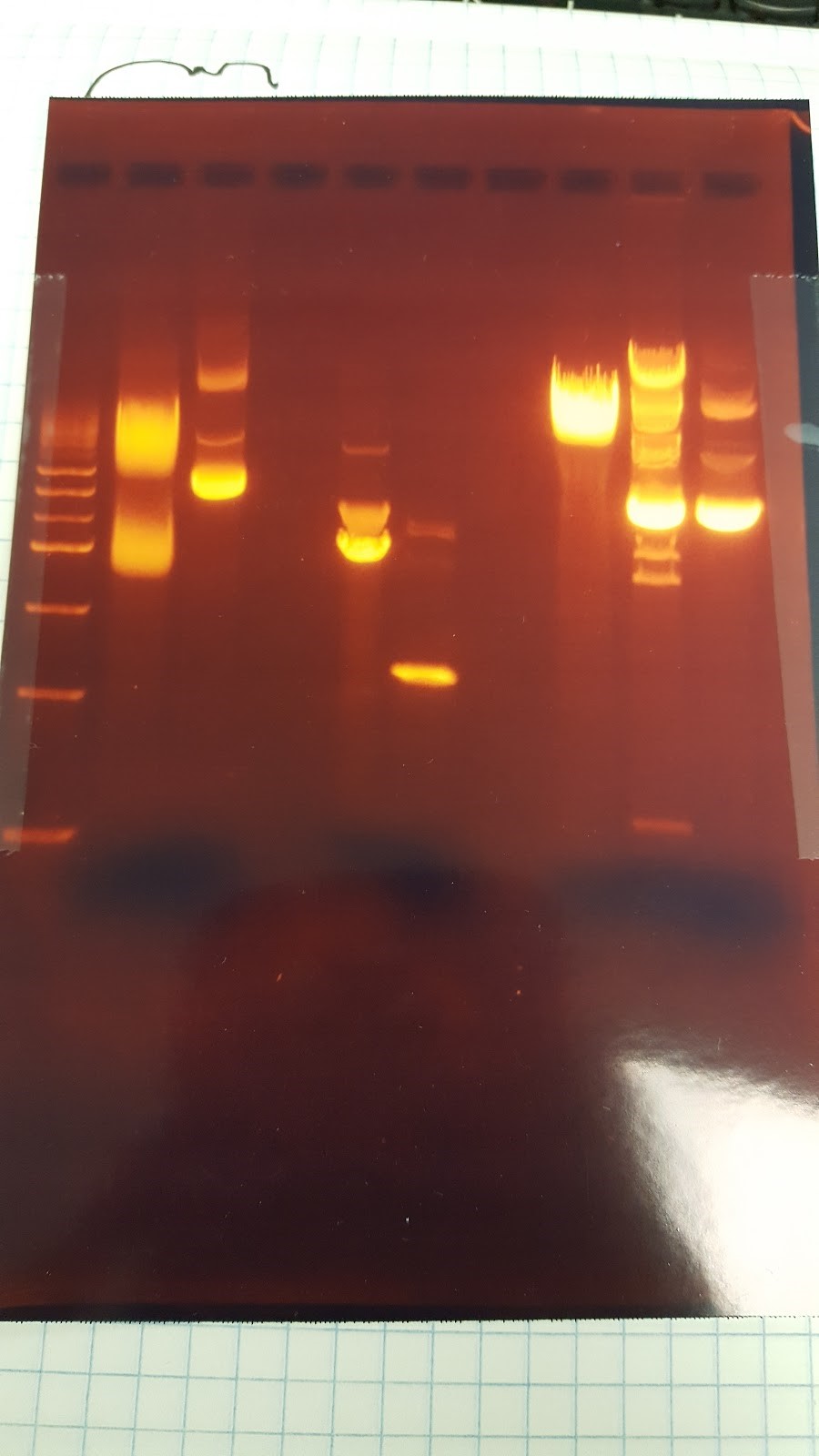
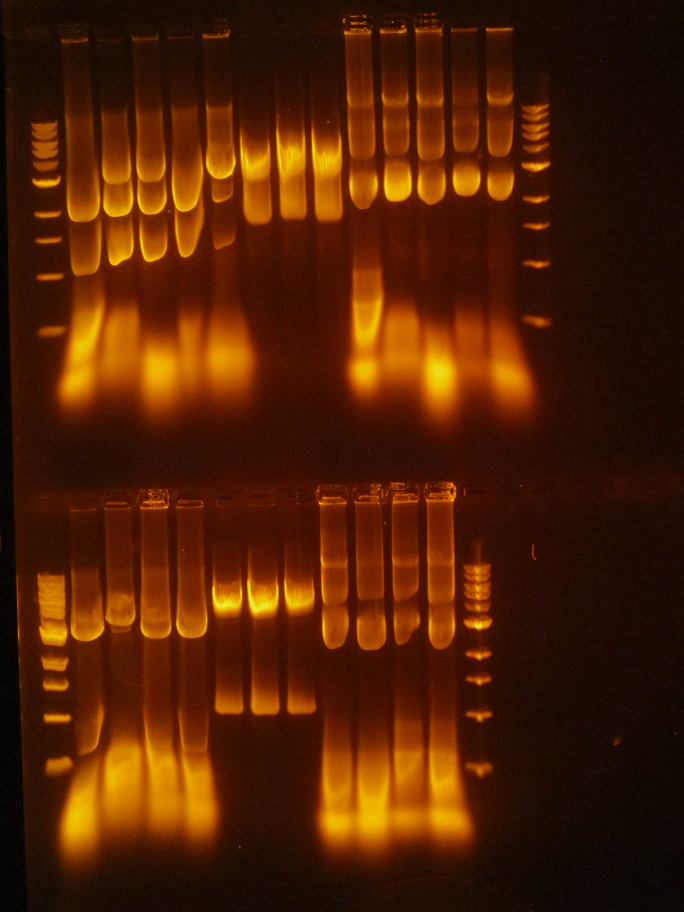
(HC) Reran the digest because I cut out the wrong bands Digested 1A, 1B, 2A, and 2B with NheI and NsiI 5 μL of DNA 1 μL of NheI 1 μL of NsiI 1 μL of Cutsmart buffer 2 μL of Sterile Water (JM/CH) The first thing we are going to do is run a digestion reaction with the enzyme Pst1 Today we ran a giant gel with 20 wells. Wells 1-10: Ladder / 416B-1 cut / 416-1 cut / Uncut 416B-1 / 416B-2 cut / 416-2 cut / 416B-2 uncut / 416B-3 cut / 416-3 cut / 416B-3 uncut Wells 11-20: 426B-1 cut / 426-1 cut / 426B-1 uncut / 426B-2 cut / 426-2 cut / 426B-2 uncut / 426B-3 cut / 426-3 cut / 426B-3 uncut / Ladder#2 Key: B- blunted and ligated DNA 1,2,3- designation of different colonies (XX) Nanodrop pSBIC3 mRPS12 mls gel extract resulted 32.9 ng/μL Goal: NEBuilder to form mls-yeGFP Transformation mls-yeGFP Redo TU digest and harvest TU Major steps: NEBuilder (on ice): 2 mls digest 8 16-4 fragment 10 hifi master Thermocycler 15 min. and store @ -20C Transformation: plate on LB-chloro TU gel extraction: pSB416 GPD EcoRI/PstI: 47.9 ng/μL (linearized plasmid) TU digest: 11.3 ng/μL7/22/16
(HC) Ran the gel of the digest from the day before 1kbp Ladder / 1A uncut / 1A cut / 1B uncut / 1B cut, 2A uncut / 2A cut / 2B uncut / 2B Cut (INSERT PICTURE HERE) Extracted from the gel lane 5 and Lane 7 Ran gel extraction from the monach gel Extraction kit. Precipitated the DNA 1. Add 1-2 μL of 5M NaCl to 30μL of DNA 2. Add 62μL of ice cold EtOH 3. Let sit on ice for 30 minutes 4. Centrifuge @ 0℃ for 10 minutes @ 13 krpm 5. Remove supernatant 6. Fill tube halfway with 70% EtOH 7. Centrifuge @ 4℃ for 2 minutes 8. Remove supernatant and allow to dry 9. Add 6μL of water Ran nano drop| Nucleic Acid Concentration | A @ 260 nm | A @ 280 nm | 260/280 | 260/230 |
|---|---|---|---|---|
| 1.5 ng/μL | 0.031 | 0.024 | 1.28 | 0.01 |
| 2.6 ng/μL | 0.052 | 0.011 | 4.53 | 0.00 |
7/23/16
(XX) Miniprep pSBIC3 mls-yeGFP: mls 1 to mls 67/24/16
(XX)Goal: Diagnostic gel to test whether mls-yeGFP has insert mls is small and will run off the gel, but mls-yeGFP can be detected; should see two bands if successfully inserted Gel Order: L, mls 1, mls 2, mls 3, Blank, A1, B1 Gel results: minipreps mls 3, A1, and B1 seems to work7/25/16
(HC)Ran NEBuilder with 1μL of DNA 1μL of Fragment 16-3 10μL of Builder Master Mix 8μL of Sterile Water Made chemically competent Cells Plated competent Cells (JM/CH) Today we are going to do boiling preps for the analytical digest today to verify if the Pst1 site has been properly cut out. The boiling preps were done with 4 tubes of 416 CYC1 and 5 tubes of 426 GPD. (XX) Goal: Continue on diagnostic gel to test mls 4, 5, and 6 Quick ligation to insert mls-yeGFP into pSB416 GPD plasmid Transformation Major steps: Mls-yeGFP digest: 5 DNA 1 EcoRI-HF, 1 PstI-HF 1 CutSmart Buffer 2 sterile water Gel order: L, mls 4, mls 5, mls 6, Blank, A2, A3, B2 Gel Results: mls 4, mls 5, mls 6 and A2 seem working!! Other activities: Gel extraction mls-yeGFP DNA from gel: mls 4 = 0.185 g mls 5 = 0.166 g mls 6 = 0.216 g Nanodrop DNA reading: mls 4: 17.8 ng/μL mls 5: 14.6 ng/μL mls 6: 16.7 ng/μL Quick Ligation: 1.5 EcoRI/PstI Cut pSB416 GPD (vector) 1 mRPS12 mls-yeGFP (insert) Follow NEB Quick ligation protocol online *Do NOT heat inactivate!! Transformation mls 4, mls 5, mls 6, A2 (transform TU miniprep back into pSB416 in order to make a stock)7/26/16
(HC)Inoculated Cultures of LB Amp with selected cells (INSERT PICTURE HERE) (JM/CH) Today is the day we find out if the blunt end ligation has worked. We are going to be running a digest and gel verification. FIRST ROW: ladder / b416 pst1 / b416 pst1 / b416 pst1 / b416 pst1 / b416 pst1 / 416 pst1 / 416 pst1 / 416 pst1 / Uncut b416 / Uncut b416 / Uncut b416 / Uncut b416 / Uncut b416 / ladder SECOND ROW: ladder / b426 pst1 / b426 pst1 / b426 pst1 / b426 pst1 / 426 pst1 / 426 pst1 / 426 pst1 / Uncut b426 / Uncut b426 / Uncut b426 / Uncut b426 / ladder Any strain with a "b" in front of it means that it has gone through blunt end ligation.(XX) Goal: Nanodrop original mls 4, mls 5, and mls 6 minipreps Inoculate culture Nanodrop readings: mls 4 = 184.2 ng/μL mls 5 = 125.4 ng/μL mls 6 = 163.6 ng/μL
7/27/16
(HC) Spinprep kit didn’t come in so I re-inoculated cultures in LB Amp (JM/CH) Today we are going to rerun the gel to try and get better results, apparently the boiling preps need to be centrifuged for ~15 min for better results. ladder / b416 pst1 / b416 pst1 / 416 pst1 / Uncut b416 / Uncut b416 / b426 pst1 / b426 pst1 /426 pst1 / Uncut b426 / Uncut b426 / ladder Any strain with a "b" in front of it means that it has gone through blunt end ligation.(XX)Goal: Dilute those DNA down to 100 ng/μL for sequencing Regrow culture Major steps: Miniprep volume left – about 24 Add 12 sterile water into A2, mls 4, mls 6 Add 6 sterile water into mls 5 Nanodrop readings after dilution: A2: 41.4 ng/μL mls 4: 94.7 ng/μL mls 5: 104.2 ng/μL mls 6: 107.3 ng/μL Sequencing primers: pSB416 GPD M13 forward (-20) [46] M13 reverse [47b] pSBIC3 VF2 [125] VR [126]