Proof of Concept
Introduction
The ultimate goal of our project is to demonstrate biofilm dispersal of a fully assembled wound dressing, consisting of a spider silk scaffold to which a variety of biofilm-degrading 'combat proteins' had been conjugated. Whereas this final test of our project remains to be performed, we have demonstrated that the key components of our SMITe system work as expected. There are three steps to our goal:
Step 1: Prove that the combat proteins possess antagonistic activity against clinically relevant bacteria. This includes demonstrating their ability to inhibit or degrade biofilms as well as certain combat proteins' bacteriolytic activities.
Step 2: Prove that Sortase A may be used to conjugate biofilm-dispersing enzymes to spider silk.
Step 3: Prove that the finished product, combat proteins conjugated to spider silk by using Sortase, is capable of disrupting biofilm.
So how did we test this?
Kirby-Bauer tests and Crystal Violet Biofilm assays have been carried out investigating step 1, a more detailed procedure of how these methods work is described below.
SDS-PAGE has been performed to analyze the conjugation reaction of Sortase A described in step 2.
Unfortunately, we have not acquired any results to confirm Step 3. However, we do believe that we have shown a proof of concept by successfully testing our two main hypothesis separately.
Kirby-Bauer
A fast procedure to characterize a compound's bacteriolytic or bacteriostatic activity. Filter papers soaked in crude cell lysates containing our combat proteins were placed on agar plates onto which TOB1 E.coli had been spread. Around the filter papers, halos with impeded bacterial growth could be observed, indicating a bacteriolytic or bacteriostatic activity of our combat proteins.
Biofilm Assay
The second method used to characterize our combat proteins was a crystal violet biofilm assay. Cultures of S. aureus or P. aeruginosa were allow to form biofilm and subjected to treatment using cell lysates containing our combat proteins. The biofilm was subsequently stained with crystal violet and the absorbance measured.
Two different effects were assessed: To determine the inhibitory effect on biofilm formation, cell lysates were added to the cultures immediately and analysed after incubation in conditions encouraging biofilm formation. To determine biofilm dispersal, cel lysates were added to established biofilms instead. Generally, lysates of untransformed cells were used as negative controls to exclude unspecific effects and gentamicin (30 μg/ml) served as a positive control.
Step 1 - Demonstrating bacteriolytic and biofilm-dispersing activity
Lysostaphin
Our combat protein lysostaphin has shown positive results with both methods and was especially effective towards S. aureus. Figure 1 shows the result from the Kirby Bauer test where halos are visible. The induced cells shows a larger halo than the non-induced cells, indicating that lysostaphin is capable of lytic activity.
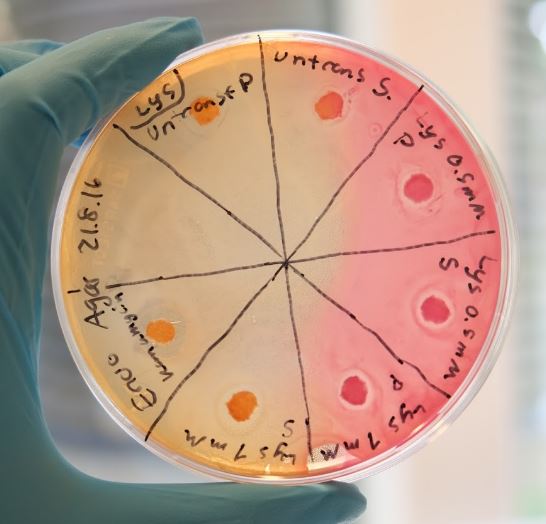
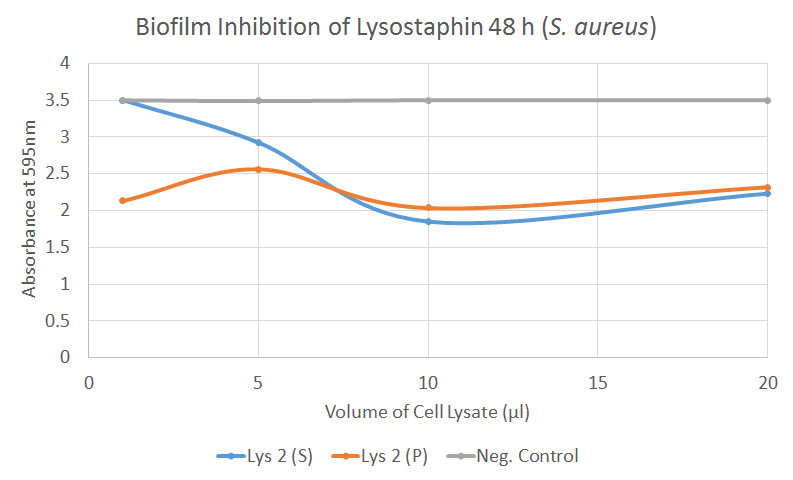
In our biofilm assays both soluble and insoluble fractions of lysostaphin-containing cell lysates displayed a clear inhibitory effect (Figure 2) Lysostaphin also effectively dispersed the pre-existing biofilm, as shown in the Figure 3 - the effect could even be observed by the naked eye. More of these results can be seen in the lysostaphin Results page.
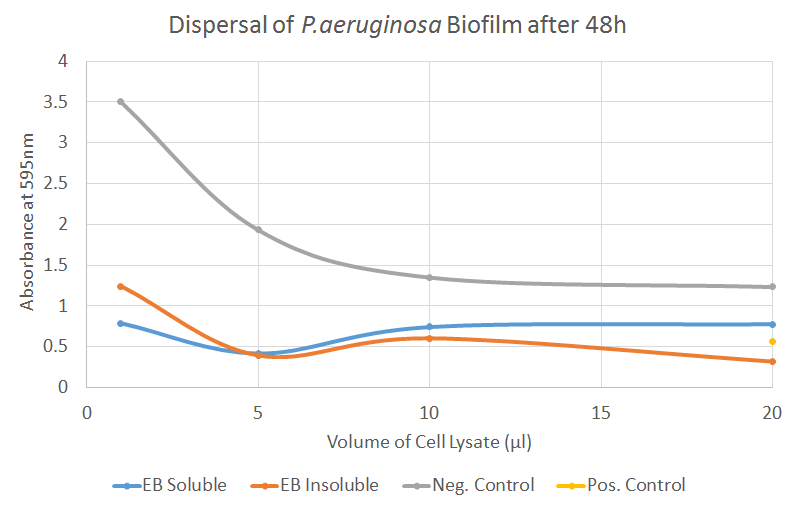
Esp
Esp (here referred to as EB, 'basic Esp') detaches biofilm by cleaving cell wall anchored proteins of S. aureus. Our synthesized EB displayed potent biofilm dispersal capacity in the Biofilm assay for both S. aureus and P. aeruginosa. For P. aeruginosa the treatment with Esp was as efficient as the positive control in Figure 5. Further results can be seen on the Results page.
Nuc
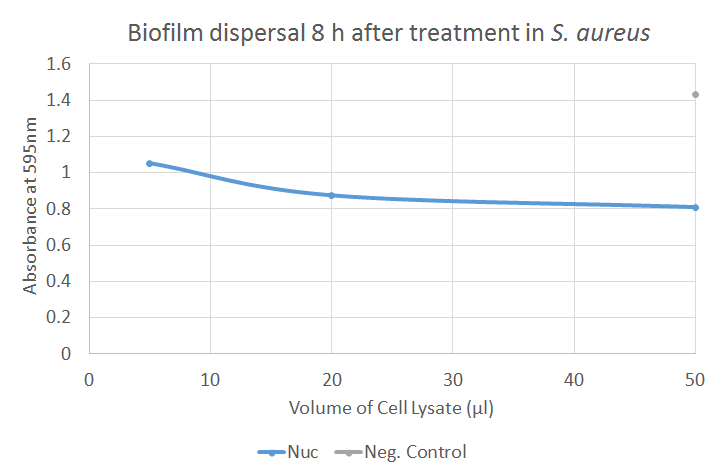
The Kirby Bauer results for Nuc were found to be negative. This was expected as Nuc targeted exposed nucleic acids which are a component of the biofilm, rather than that of free living cells. In the biofilm dispersal assay, Nuc showed good activity; in Figure 6, a strongly decreased absorbance could be observed after application of crude cell lysates containing Nuc, indicating a clear dispersal of the biofilm. Further results can be seen on the Results page.
Defensin
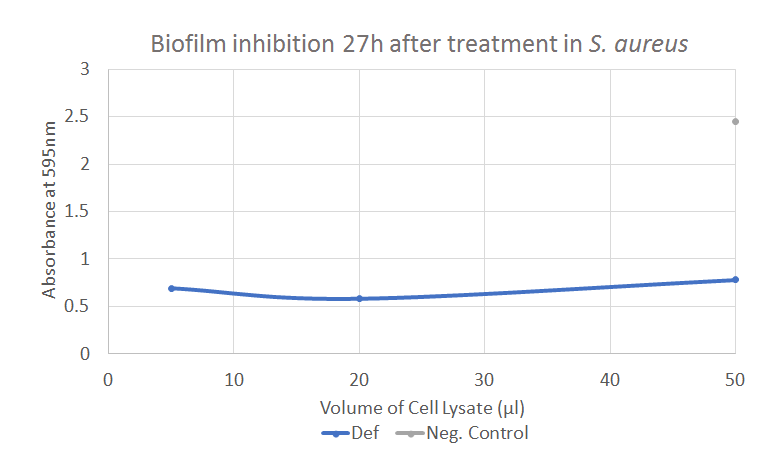
Defensin has showed positive results in both the Kirby-Bauer tests and the biofilm assay. The result from the biofilm assay is displayed in Figure 7, showing a significant decline in the absorbance compared to the negative control. Further experiments and results can be seen in the Results page.
Step 2 - Sortase-mediated conjugation
In this section we have proven that Sortase works as a catalyzer of a conjugation reaction between two fusion partners with a LPETGG-tag and a exposed glycine residue. The spider silk as well as the Sortase and tagged peptide (ZHER2) were kindly given to us by our supervisors and mentors, My Hedhammar and Kristina Westerlund, respectively.
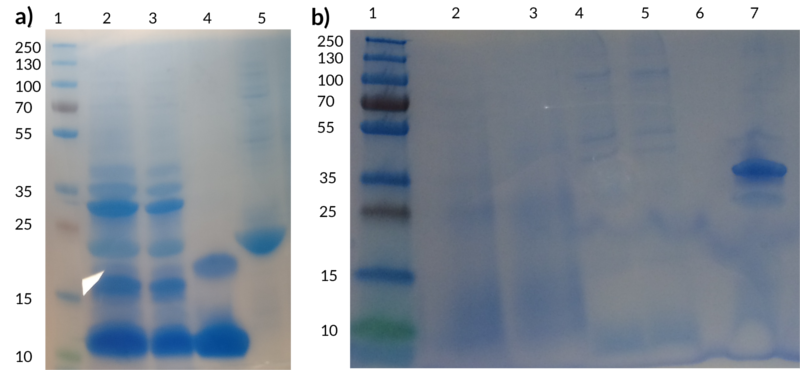
The figure below shows two SDS-PAGEs performed to demonstrate the efficient conjugation of peptides to spider silk. Figure 7(a) shows a test performed with spider silk and ZHER2. The bands visible between 30-45 kDa in well 2 and 3 correspond to the conjugated product, demonstrating clearly that Sortase is attaching the tagged peptide to the spider silk.
In the course of our project, we were also able to create additional BioBricks containing a combat protein and what we refer to as 'LT' - a sequence consisting of a linker, a Sortase recognition motif LPETGG as well as a Hisx6 tag for purification. We were able to demonstrate efficient attachment of our defensin BioBrick to spider silk as well, proving that the design of our BioBricks permit Sortase-mediated conjugation.
In Figure 7(b), the conjugated product can be seen observed in well 7 around the size 35 kDa, corresponding to the size of spider silk (~23 kDa) and defensin (~11kDa) fused together. The negative controls are unfortunately not visible because too little protein was left to be loaded onto the gel but the same control (unconjugated spider silk) can be seen in well 5 of Figure 7(a). In the lab books for defensin week 16 the Sortase-A-mediated conjugation between spider silk and purified defensin is described in more detail.
A Look Beyond
Based on the results, several combat proteins have shown efficient biofilm dispersal or inhibition. We have further shown that a conjugation between a protein and our BioBrick defensin using Sortase is fast and efficient.
Demonstrating that our final wound dressing, consisting of combat proteins conjugated to spider silk, is capable of degrading biofilm still remains as future work. With more time in the lab, we would hopefully have been able to analyse the device, defensin conjugated to spider silk, to investigate its biofilm dispersing ability.
Nonetheless, we have seen speedy progression and several positive results with defensin. Notably, we were able to demonstrate successful degradation of both S. aureus and P. aeruginosa biofilms and were able to conjugate the purified defensin to spider silk using a commercial Sortase A. Thus, the two main goals of our project - biofilm degradation and Sortase-mediated attachment to spider silk - have successfully been demonstrated, providing a solid base for any future on SMITe - Spider silk Mediated Infection Treatment.


