Sortase A
Summary
Sortase A has the potential of becoming an essential part within the iGEM community. We thus aimed to create a Sortase A BioBrick making it available for future iGEM teams to utilize for conjugation and immobilization of proteins.
DNA extraction of S.aureus genome [✔️]
Isolation of truncated Sortase A [✔️]
Gibson assembly of GB1, Sortase A and pSB1C3 [❌]
Transformation of Gibson assembled product [❌]
Overlap extension PCR GB1 & Sortase A [✔️]
Digestion, Ligation and Transformation of Overlap Extension PCR Product [✔️]
Site directed mutagenesis for removal of XbaI restriction site [❌]
3A Assembly of Sortase A Biobrick, T7 promoter and pSB1C3 BioBrick [❌]
Expression and characterization of Sortase A BioBrick [❌]
Results
DNA extraction of S.aureus genome
The initial step toward creating the Sortase A BioBrick aimed to extract the S.aureus genome from Staphylococcus aureus Newman strain using a standard genomic DNA extraction kit.
Isolation, First Attempt [✔️]
Based on gel electrophoresis and DNA concentration, the S.aureus genome was successfully isolated. Subsequent step aimed to isolate truncated Sortase A gene, i.e. without its membrane anchoring domain, using suitable designed primers.
Isolation of truncated Sortase A
By designing suitable primers, specifically a forward primer annealing 59 amino acids downstream the coding sequence for Sortase A, we obtained a truncated version of the gene missing the membrane anchoring coding domain and GB1 sequence overhang to prepare for Gibson assembly.
Truncation, First Attempt [✔️]
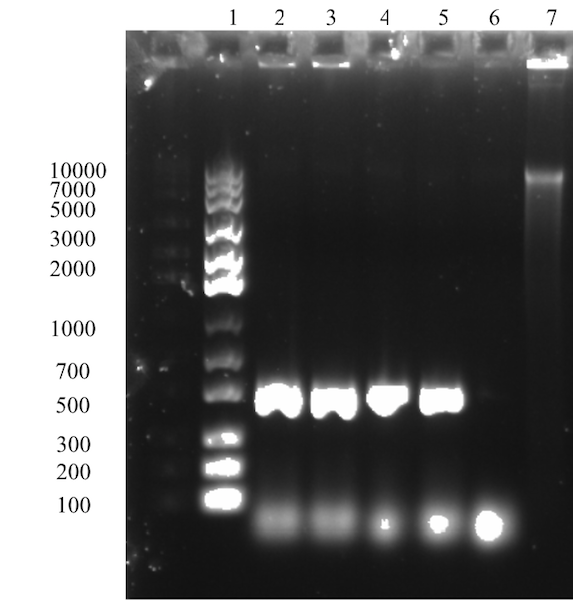
The expected size of the Sortase gene at 491 bp is observed for all four samples in the gel (Figure 1). Further, no other unspecific bands are observed on the gel and the negative control shows no sign of amplification. In conclusion, isolation of truncated Sortase A gene was successful. Next step aims to assemble the gene Sortase gene fragment with the solubility tag GB1.
Gibson assembly of GB1, Sortase A and pSB1C3
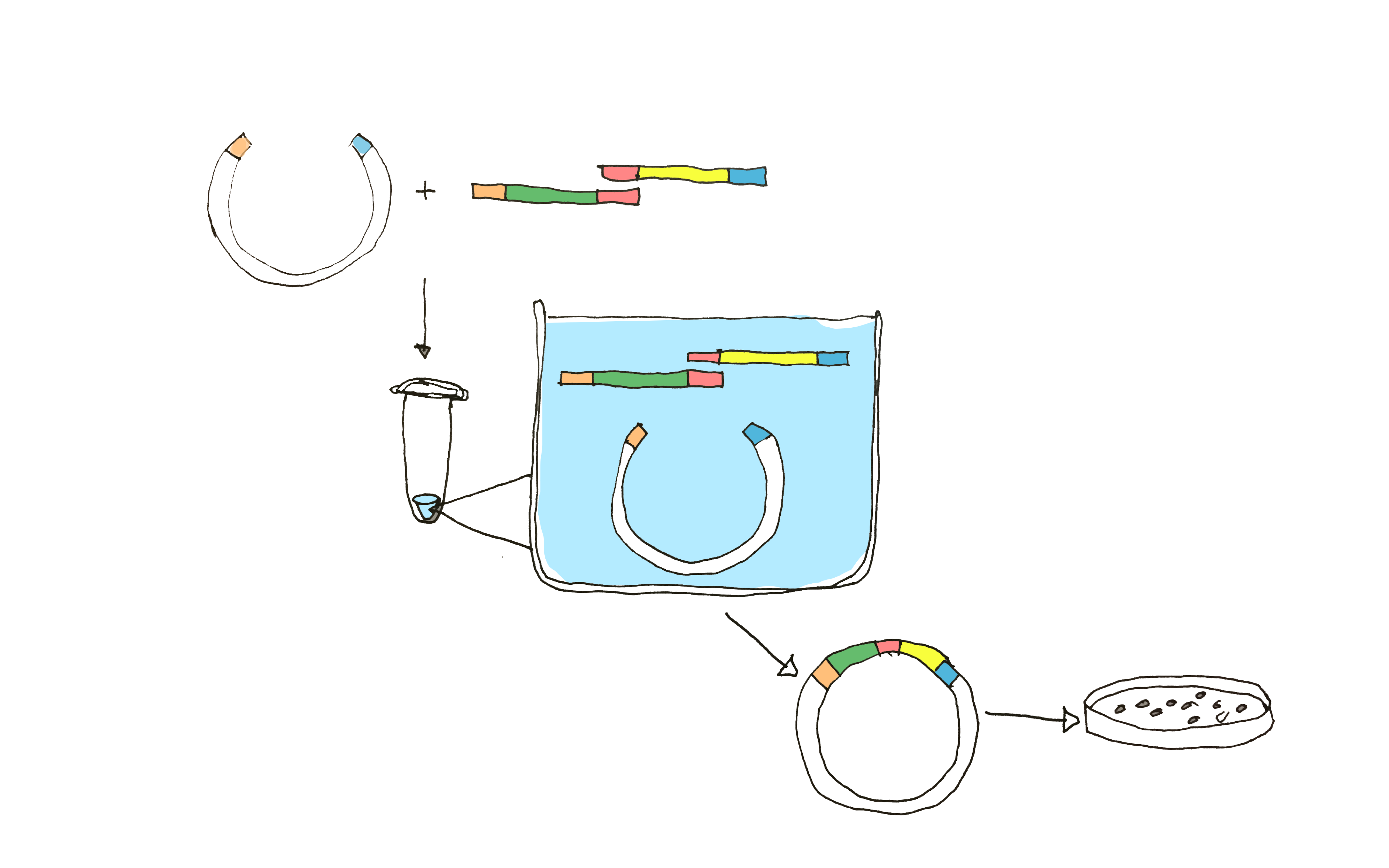
Using NEB Gibson assembly Master Mix, we aimed to assemble truncated Sortase A with the solubility tag GB1. In the assembly mix, a 5’ exonuclease chews back the 5´ end sequences generating overhangs designed with complementary sequences to the fragments to be annealed with. DNA polymerase fills in the gaps of the annealed single strand regions, where the nicks are finally sealed with a DNA ligase (Figure 2).
Gibson Assembly, First Attempt [❌]
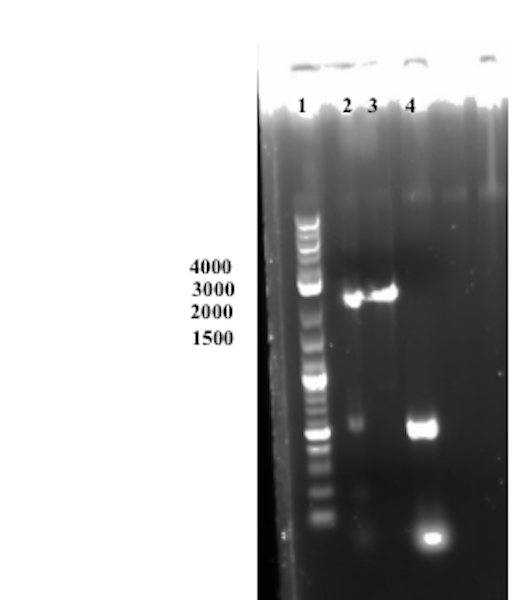
Well 2 was loaded with the Gibson assembly sample. The expected size of the assembled product is around 3000 bp, which cannot be observed in the gel below. An undiluted sample of Sortase A was used in the experimental set-up thus the molar ratios between the different fragments to be assembled was skewed. In conclusion the assembly was deemed unsuccessful.
Gibson Assembly, Second Attempt [❌]
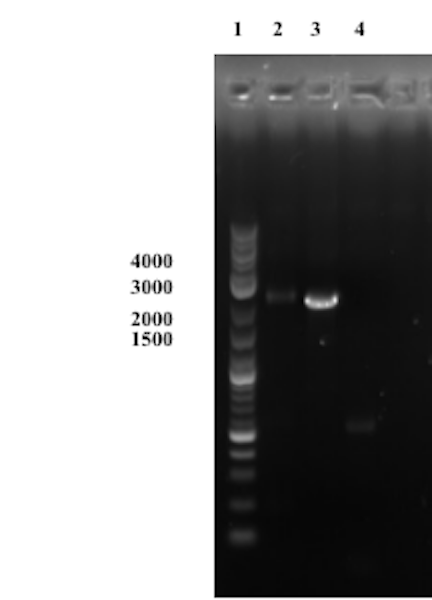
Compared to the first trial, no bands except one faint band approximately at the right size of the presumably assembled product are observable in well 1. Though this could indicate on a successful Gibson assembly we consider the results inconclusive until a successful transformation and sequencing of the product has been conducted.
Transformation of Gibson assembled product
Following Gibson assembly, transformation of samples from both trials was conducted, to acquire colonies for plasmid purification, sequencing and preparation of site-directed mutagenesis.
Transformation, First Attempt [❌]
5 μl of Gibson assembled product was used for transformation into TOP10 E.coli. The transformation generated no colonies after overnight incubation. Transformation was thus unsuccessful and was re-attempted with more DNA.
Transformation, Second Attempt [❌]
10 μl of Gibson assembled product was used for transformation into TOP10 E.coli, increasing the amount two-fold from Trial 1. The transformation generated no colonies after overnight incubation. Transformation was thus unsuccessful. A new strategy was planned, using overlap extension PCR to join the two fragments together before cloning into pSB1C3 vector.
Overlap extension PCR GB1 and Sortase A
By using PCR fragments of Sortase A with a 20 bp sequence Homology to GB1 we aimed to perform an overlap extension PCR to join the two fragments together. Two different annealing temperatures (66°C & 69°C) were tested to optimize primer binding.
Overlap Extension PCR, First Attempt [✔️]
Bands at around the expected size (677 bp) of the joint fragments can be observed for all samples in the first gel below (Figure 5), although they are significantly stronger for samples treated with an annealing temperature at 66°C, thus these samples were chosen to be gel purified given that there still are unspecific bands observed on the gel, which might be due to weaknesses in the primer design.
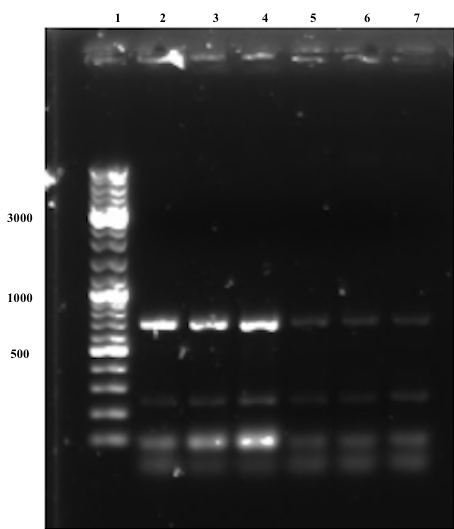
The second electrophoresis was performed after gel purification (Figure 6), where no unspecific band are observed. In conclusion, the overlap extension PCR was successful. Subsequent step aimed to digest the fragments with EcoRI and PstI to ligate it into pSB1C3 BackBone.
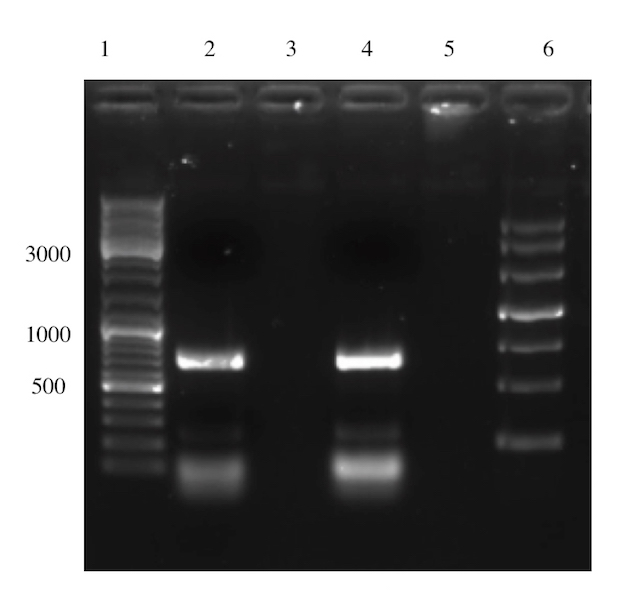
Digestion, Ligation and Transformation of Overlap Extension PCR Product
Digestion of EcoRI and PstI sites on Overlap extension PCR product was performed with the goal to further ligate the fragment into an equivalently digested and linearized pSB1C3 vector. Transformation of Overlap extension PCR product, ligated into pSB1C3, in TOP 10 and BL21 chemically competent cells.
Assembly and Transformation, First Attempt [✔️]
Two colonies were observed one on the BL21 plate and another on the TOP10 Plate. Both samples were picked and inoculated for plasmid extraction. The purified samples had a final concentration of 65.8 ng/μl and 117.4 ng/μl respectively.
Site directed mutagenesis for removal of XbaI restriction site
Sortase A contains an XbaI restriction site in its sequence which, by iGEM rules, can not be present in a BioBrick. Therefore, site directed mutagenesis (SDM) was attempt to remove it.
Two sets of primers were designed, Back to Back primers (BBP) and overlapping primers (OLP). The primers were designed to anneal with 20 base pairs at the location of the site, with one base pair switched at the region annealing to the restriction site. Template DNA after PCR would be taken care of with DpnI and the nicks sealed with ligase.
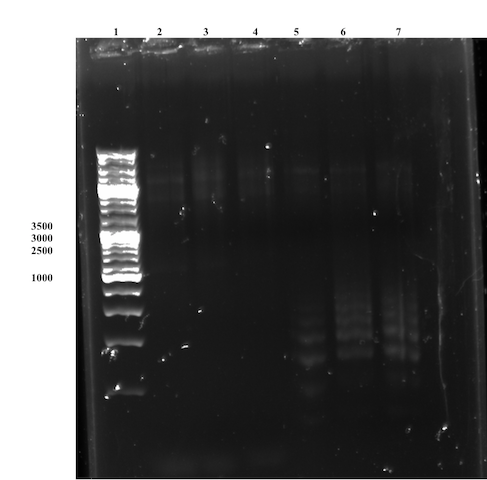
SDM, First Attempt [❌]
Observing the gel, the conclusion was quickly drawn that the Site directed mutagenesis was unsuccessful for both primer pairs. No strong Bands of interest are observed on the gel (Figure 7), though unspecific bands can be observed for the overlap primer-set. The failed attempt is most likely caused by lacking primer design, which needs to be reviewed and possibly changed substantially, e.g. design primer pairs with an higher Tm to avoid unspecific binding.
Future outlook
In general, a majority of the steps towards a finished BioBrick has been performed. The assembly of the finished vector was an iterative process in which the first approach performing a Gibson Assembly were not as straightforward as initially expected. Shifting the molar ratios and total molar amount of fragments for an optimized assembly would have been favorable.
Further, the downstream process development of the BioBrick could have been reduced with one step. The customary synthesized GB1 solubility tag could have been designed with a T7 promoter upstream the coding sequence, making the 3A assembly step obsolete.
In conclusion, the development of the Sortase BioBrick is not far from being realized. There are a few critical steps left to be performed, which could be a suitable subproject for future iGEM teams to take on, given the broad range of applications Sortase A could be utilizing for.


