Safety
One of our major goals was to not only conduct the lab work safely but also think beyond our project up to the clinical application of our wound patch. Therefore, we combined the clinical expertise of our team members with our knowledge of biotechnology to design a project that was not only innovative but also specifically designed to be as safe as possible for clinical applications.
Live Bacteria in Finished Product
Chronic Wounds are considered to be non-healing because of two major factors: wound infection and a non-physiological, ineffective inflammatory response [1].
With an already infected wound and likely impaired immune response we were hesitant to create a ‘living’ bandage i.e. one which contained bacteria themselves, which could potentially aggravate the chronic wounds in two ways. Firstly, multi-species biofilms have been shown to be symbiotic, e.g. by ‘passive resistance’ when the resistance of one organism protects another strain or species which on its own would be susceptible to the therapeutic agent. This process in particular could be detrimental when live bacteria are incorporated into a wound dressing - the bacteria would likely have resistance genes used for selection and could therefore potentially protect the bacteria infecting and colonising the wound [2]. Secondly, even just cellular debris of bacteria can be strongly immunogenic and might further interfere with the inflammatory response, which has already been highly altered in chronic wounds. In particular the Toll-like Receptor 4, part of our microbe detection system, reacts strongly to bacterial membrane components and has been shown to inhibit healing of chronic wounds [1] [3].
Spread of the therapeutic substances
In our decision-making process, we originally wanted to focus on staphylococcal infections and biofilms formation on implanted medical devices, which share many of the issues of chronic wound infections [4]. One of the major concerns with this project proposal was that in order for it to be applicable to real-life situations, the proteins added to the implants would have to be kept from seeping into the surrounding tissue and blood stream to avoid potentially grave, or at least unknown, secondary effects. This was one of the reasons we settled on our SMITe project.
With SMITe, the same concern exists but is comparably smaller - the thick layer of biofilm will effectively shield the healthy tissue from the action of the enzymes and the fact that circulation is normally impeded in chronic wounds decrease the risk of spreading our combat proteins to healthy tissue [5].
Additionally, the fixation of our combat proteins to the spider silk via Sortase A is based on the formation of a peptide bond. This covalent linkage is very stable and should withstand the conditions in the chronic wound without undergoing degradation [6]. In one of our own experiments, it can be seen that even the heating process and reducing conditions of an SDS-Page sample preparation did not detach the peptides from the spider silk.
Unspecific Effects
Unfortunately, designing a treatment without side effects is close to unachievable but measures can be taken to minimise them. In our case, unspecific action of two of our combat enzymes is the major issue in terms of side effects. One of those enzymes, Nuc, does not show strong specificity for either eukaryotic or prokaryotic DNA, indicating that it might not just degrade extracellular bacterial DNA but also Neutrophil Extracellular Traps (NETs), DNA secreted by Neutrophils that constitutes a major part of the immune defense in wounds [7] [8].
In order to avoid those unspecific effects, we considered several alternative enzymes for SMITe which would show specificity for bacterial DNA, based on its different methylation pattern. There are interesting candidates with such specificities such as SmaI but due to time constraints we decided to focus on Nuc for our SMITe project [9]. If SMITe or a similar concept would be adapted for use in the clinics, the role of neutrophils in chronic wounds would have to be further evaluated since it is unclear at this point whether they overall aid or further disturb the healing process [1]. If neutrophils were found to contribute significantly to wound healing and pathogen defense during the stages our dressing is intended to be applied, more measures would have to be taken. One of those could be attaching Nuc only at specific parts of the dressing where the biofilm has a considerable thickness and Nuc would be unlikely to reach NETs.
Esp is a second enzyme that might show undesirable effects since it is a serine protease with not entirely clear specificity [10]. However, Esp is usually secreted by S.epidermidis colonising the skin and mucous membranes of many healthy humans and to our knowledge has not been documented as causing any kind of ill effect [11]. Thorough safety testing would of course be needed if SMITe was ever going to be used in the clinics and, similarly to Nuc, attachment only to limited parts of the wound dressing could be considered.

A futuristic imagining!
These concerns feed into a conceptual image for the future of SMITe, when chronic wounds could be mapped and their individual biofilms characterised through imaging, for example through confocal microscopy. If it were ever possible to achieve this we imagined SMITe as a product which could then be designed to reflect the structure of individual wounds themselves; targeting thicker and thinner areas of the biofilm with specifically distributed proteins, and areas where colonisation or infection was most dominated by certain microorganisms with appropriate dispersal agents. This would be a entirely new avenue in personalised medicine products and is one way this field could spread across disciplines and give a whole new meaning to getting a dressing which “fits".
Release and Dispersal of Bacteria
If the presence of bacterial biofilm is a barrier to healing, then it follows that dispersal and degradation of the biofilm structure would facilitate wound healing. The two most important remaining questions in this case would be: what happens to the remnants of the biofilm once degraded (and the SMITe, if it breaks down) and also, whether the release of dormant bacteria residing within the biofilm generates bacteremia in the patient? Is the approach we have chosen worth the risks of further spreading the infection?
Considering the latter, there is a theoretical possibility that once released from the chronic wound via the biofilm being degraded, bacteria become susceptible to antibiotic treatments which were previously ineffective as the drugs were formerly unable to penetrate the biofilm structure. Another possibility is that once the biofilm has been degraded and the human immune system encounters the ‘debris’ in the bloodstream, an immune response would be initiated which is able to eradicate the infected or colonised tissue altogether. This would avoid applying SMITe alongside ‘antibiotic cover’, which would be the ideal situation from the point of view of reducing antibiotic resistance. In any case, patients would need to be carefully monitored to detect indications of systemic inflammatory response syndrome and consequent sepsis until progress in wound healing was apparent.
In accordance with many aspects of our project, meticulously monitored clinical trials would be essential in establishing the safety of SMITe… and we are currently very distant from these being realised.
Contributing to patients’ health and safety
The ultimate goal we had for SMITe was contributing to the safety and improved life-quality for chronic wound patients, which would only be possible if SMITe was optimally adapted to the situations most often encountered in the clinics.
A great source of discussion in the project planning stage of our work was the timing and stage of biofilm development for which SMITe was designed. We anticipated that to enable SMITe to have optimum beneficial effects, the dynamic condition of the biofilm needed to be considered and this matched to some practical issues. One of the many logistical problems of chronic wounds is that by definition they are well established at the point of diagnosis. Since these are defined as having not healed for a prolonged period (usually weeks), [12] we had to anticipate battling an established biofilm; a trickier task altogether.
We considered that if we instead wanted to aim for preventing the development of chronic wounds we would need to also devise a system for risk stratifying acute injuries in order to treat those most at risk in anticipation. This seemed an unrealistic endeavour for our project but does continue to highlight the fact that meticulous screening of elderly individual’s pressure points, acute injuries, strict monitoring of diabetics and those with vascular disease is essential, [12] as well as education about the complexities of managing these injuries.
Overall, it became evident that our project would be unlikely to protect vulnerable patients from suffering high risk injuries predisposing them to chronic wounds and we agreed that our ability to intervene in the early stages of wound development was limited by the patient demographic; these individuals often present to healthcare establishments with an established wound.
It was for these reasons that SMITe was designed to disperse established biofilms which we consider to be the most realistic approach for clinical applications and which could considerably contribute to chronic wound patients’ safety and well-being.
Minimising Antibiotic Resistance
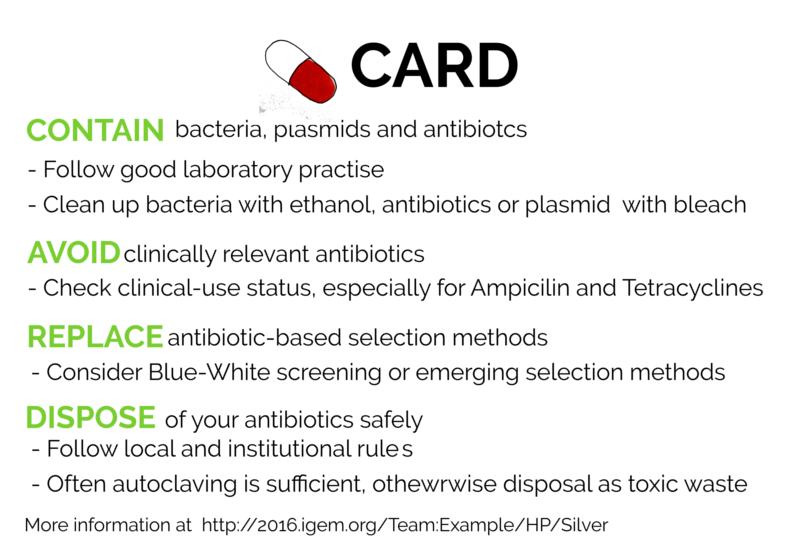
A big motivation for conducting SMITe was also antibiotic resistance and how it impacts both our presence and future. As part of our Human Practices, we collected information regarding the use of antibiotics in research and put together a set of easy-to-follow instructions and guidelines regarding the safe use of antibiotics and antibiotic-resistant organisms in research. The Project, CARD, is based on four simple principles:
Contain your resistant bacteria, plasmids and antibiotics
Avoid using clinically relevant antibiotics
Replace antibiotic-based selection methods
Discard of antibiotics safely
We designed a physical card, shown below, that can be used as a reference for students, biohackers and researchers. If you'd like to learn more about our CARD project, go to our Human Practices page
References
1: Zhao, G. et al. Biofilms and Inflammation in Chronic Wounds. Adv. wound care 2, 389–399 (2013)
2: Wolcott, R., Costerton, J. W., Raoult, D. & Cutler, S. J. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 19, 107–112 (2013).
3: Murphy, K. Janeway’s Immunology, 8th Edition. (Garland Science, 2012).
4: Lister, J. L. & Horswill, A. R. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4, 178 (2014)
5: http://www.woundcarecenters.org/article/wound-basics/chronic-wound-basics (retrieved 08.10.2016)
6: Vollhardt, Peter. Organic Chemistry Structure and Function. 6th Ed., 5th. New York : W. H. Freeman and Company, 2011
7: Okshevsky M, Regina VR, Meyer RL. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol. 2015 Jun;33:73-80
8: Murphy, K. Janeway’s Immunology, 8th Edition. (Garland Science, 2012).
9: Argudín, M. A., Rodicio, M. R. & Guerra, B. The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Lett. Appl. Microbiol. 50, 127–130 (2010).
10: Sugimoto, S. et al. Staphylococcus epidermidis Esp degrades specific proteins associatedwith staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 195, 1645–1655 (2013).
11: Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346 –349.
12: Siddiqui, A.R., Bernstein, J.M. Chronic wound infection: Facts and controversies. Clin. Dermatol. 28 519-526 (2010).


