
iGEM Tel-Hai 2016
Notebook
30.6.16
Extracting the plasmids from the iGEM kit:
- pSB1C3 - the backbone through which the parts should be sent
- J04450 - the plasmid pSB1C3 with an added RFP gene (coding a red fluorescence protein)
- K60800 - a plasmid containing a GFP gene
- All plasmids were transformed into Top 10 E. Coli strain, using the heat shock method.
- E. coli were spread onto LB agar plates with the antibiotic chloramphenicol and were incubated ON at 37o.
6.7.16
- LB was inoculated with colonies of E.coli each transformed with one of the plasmids above that grew ON
- Colonies were incubated at 37o ON
7.7.16
- Extraction of plasmids from colonies using Miniprep kit and validation of presence of plasmid by running extraction product through agarose gel (1%) - fig 1.
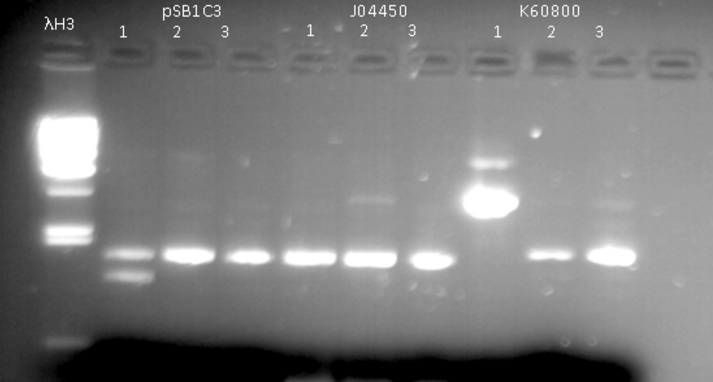
Fig. 1 - Extraction of pSB1C3, J04450, K60800
13.7.16
- Restriction of the plasmids pSB1C3 and J04450 with EcoRI and PstI. Incubation for 1 hour at 37°C
- We ran the restriction products in agarose gel (1%)
- Gel results indicated that that the plasmid J04450 was ok. With the plasmid pSB1C3 we did the procedure once again (fig 2)
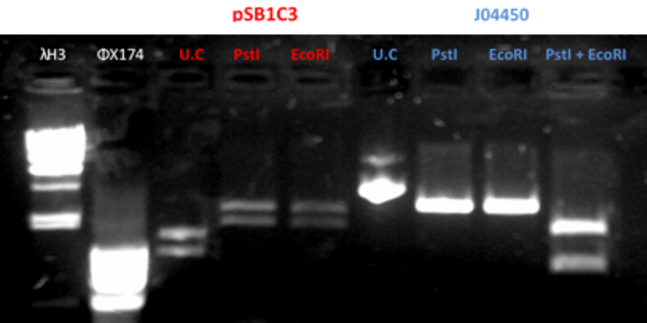
Fig.2 - Restriction of the plasmids pSB1C3 and J04450 with EcoRI and PstI
3.8.16
- We received the parts 1-4 from IDT
- part 1- contains the sequence of tGFP gene under EF1a promoter
- part 2- contains the sequence of CFTR exon 11, donor part
- part 3- contains the sequence of gRNA1 for CFTR ΔF508
- part 4- contains the sequence of gRNA2 for CFTR ΔF508
- Restriction of the parts and the plasmid pSB1C3
- Ligation of the parts to the plasmid backbone pSB1C3
- All ligated plasmids were transformed into Top 10 E. Coli strain, using the heat shock method.
- E. coli were spread onto LB agar plates with the antibiotic chloramphenicol and were incubated ON at 37o
7.8.16
- Extraction of plasmids from colonies that grew using Miniprep kit
8.8.16
- Restriction of the plasmids pSB1C3 + parts 1-4 with EcoRI and PstI. Incubation for 1 hour at 37°C
- We ran the restriction products in agarose gel (1%)
- Gel results indicated that the ligation of the plasmid with part 4 was ok (fig 3)
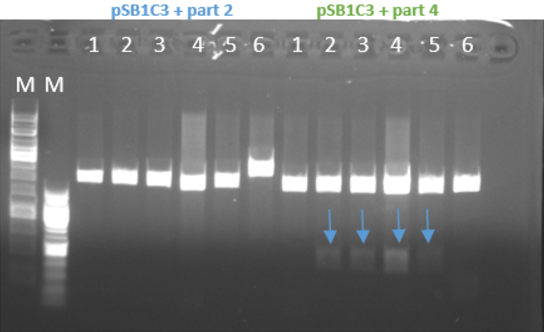
Fig.3 - Restriction of the plasmids pSB1C3 + parts 1+4 with EcoRI and PstI - With all the other parts, we did the procedure again at 10-16.8.16 and the ligation worked for part 1 and part 3 and for part 2 it didn’t work (fig 4)
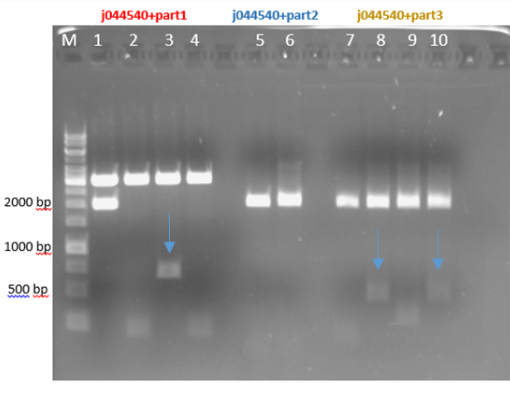
Fig.4 - Restriction of the plasmids j044540 + parts 1+2+3 with EcoRI and PstI
17-18.8.16
- We received the purified CTB protein from SIGMA-ALDRICH
- The CTB was crossed linked to the plasmid pSB1C3 using a chemical linker
29.8.16
- We received lung epithelial cells NCI-H 1650
- The cells were thawed and seeded in a 6 well plate and incubated in a suitable incubator at 37o and 5% CO2
31.8.16
- We ran the crosslink product in agarose gel (3 %)
- The gel results indicates that there is a gel shift. The DNA runs through the gel in a different way compare to the negative control (pSB1C3).
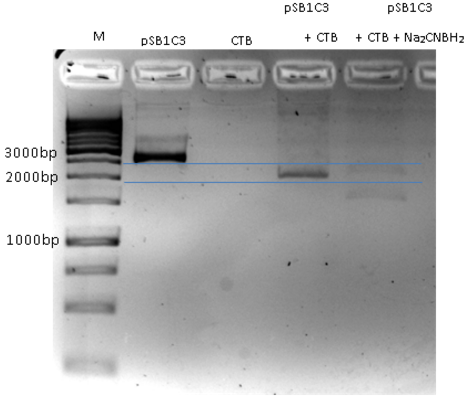
Fig.5 - crosslink product. pSB1C3+CTB migration through agarose gel 3%. First well is the marker, Second well is only pSB1C3, Third well CTB only (for negative control), Fourth Well is pSB1C3+ CTB cross link showing a gel shift due to crosslinking.
11.9.16
- we received a chimeric LTB protein with a DNA binding domain, dyed with FITC (green fluorescence)
- we ran the protein in a polyacrylamide gel for verification of the protein size
- the gel result indicate that the protein is at the correct size, ~60kD (fig 6)
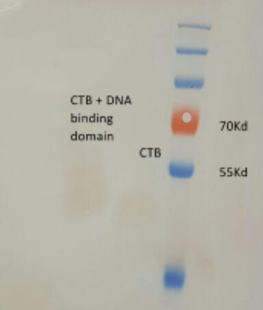
Fig.6 - LTB+DBD dyed with FITC, in polyacrylamide gel.
20.9.16
- we dyed a plasmid containing a GFP (pEGFPN3) with Hoechst stain (blue fluorescence)
pEGFPN3 plasmid - we purified the plasmid from the remaining dye using a Miniprep kit
- we incubated the plasmid + Hoechst with the LTBD + FITC for 45 min. at 37o
- NCI-H 1650 cells were washed and prepared for incubation with plasmid and protein complex and prepared for FACS
- FACS analysis below (fig 7) shows that the protein the cells but we were not able to detect the blue Hoechst staining
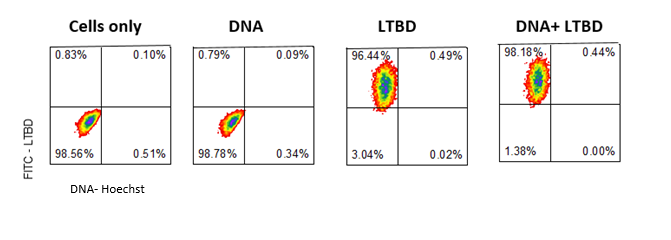
Fig.7 - FACS results
Preparing cells for confocal-
- We seeded cells in 24 well confocal plates for the experiment the next day
21.9.16
- we incubated the plasmid + Hoechst with the CTBD + FITC for 1 hour at 37o
- Cell medium was added to the complex to achieve 200µl and the whole reaction was added to the cells.
- Cells were incubated in a suitable incubator at 37o and 5% CO2 wrapped in aluminum foil for 5 hours
- Cells were pictured using a confocal microscope
- The picture shows (fig 8) the green staining of FITC inside the cells. Looking at the Hoechst staining we received a lot of background and assumed that the DNA degraded
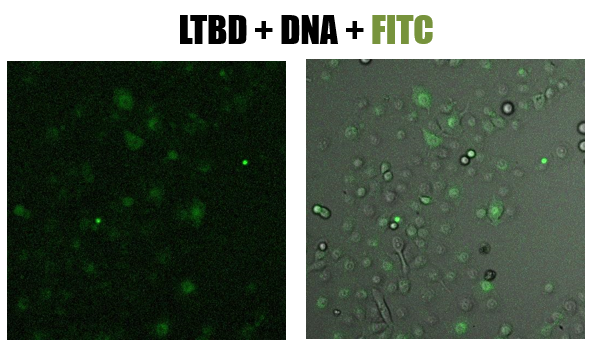
Fig.8 - confucal results. Green staining of epithelial cells (NCI-H1650) by LTBD stained with FITC. LTBD binds specifically GM1 on epithelial cells and enters by endocytosis
9.10.16
Binding plasmid in constant concentration (400ng) to CTB + DNA binding domain
- we linearized the plasmid pEGFPN3 using EcoR1, 1 hour at 37o
- we incubated the linearized plasmid with increased concentrations of the protein- 30,60,90,150,300,600 ng, and with a CTB protein without a DNA binding domain, for 45 min. at 37o
- we ran the reactions in agarose gel (1%)
- gel results (fig 9) indicate that the optimal concentration of protein that binds 400ng of DNA is 300ng
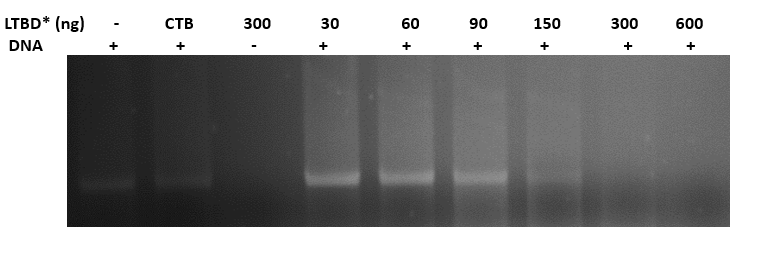
Fig.9 - Binding pSB1C3 plasmid in constant concentration (400ng) to LTBD. gel results indicate that the optimal concentration of protein that binds 400ng of DNA is 300ng. *LTBD - Heat-Labile Toxin (LT), an analog to CT

