m |
|||
| (25 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| − | + | {{:Team:Aix-Marseille/Template-Style-and-Menu}}{{:Team:Aix-Marseille/Template-Notebook}}<div class="events"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <div class="events"> | + | |
<div id="2016-06-13" class="evday"> | <div id="2016-06-13" class="evday"> | ||
| − | + | === Monday, June 13th === | |
* Preparation of heat competent DH5α cells ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol #1]). | * Preparation of heat competent DH5α cells ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol #1]). | ||
| Line 223: | Line 9: | ||
<div id="2016-06-14" class="evday"> | <div id="2016-06-14" class="evday"> | ||
| − | + | === Tuesday, June 14th === | |
* Transformation of DH5α cells with test plasmid ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation of DH5α cells with test plasmid ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 229: | Line 15: | ||
<div id="2016-06-15" class="evday"> | <div id="2016-06-15" class="evday"> | ||
| − | + | === Wednesday, June 15th === | |
* Preparation of heat competent Tg1 cells ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). | * Preparation of heat competent Tg1 cells ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). | ||
| Line 236: | Line 22: | ||
<div id="2016-06-16" class="evday"> | <div id="2016-06-16" class="evday"> | ||
| − | + | === Thursday, June 16th === | |
* Transformation of DH5α cells with pSB1C3 + RFP ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation of DH5α cells with pSB1C3 + RFP ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 243: | Line 29: | ||
<div id="2016-06-17" class="evday"> | <div id="2016-06-17" class="evday"> | ||
| − | + | === Friday, June 17th === | |
* Plasmids purification K880005, K823017, I0500, K1404006, B0034 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmids purification K880005, K823017, I0500, K1404006, B0034 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 250: | Line 36: | ||
<div id="2016-06-20" class="evday"> | <div id="2016-06-20" class="evday"> | ||
| − | + | === Monday, June 20th === | |
* pSB1C3 digestion from 17/06 plasmids purification ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Digestion_protocol_BioBrick_Assembly_Kit Protocol#4]). | * pSB1C3 digestion from 17/06 plasmids purification ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Digestion_protocol_BioBrick_Assembly_Kit Protocol#4]). | ||
| Line 259: | Line 45: | ||
<div id="2016-06-21" class="evday"> | <div id="2016-06-21" class="evday"> | ||
| − | + | === Tuesday, June 21th === | |
* pSB1C3 digestion and gel migration, NucleoSpin gel extract and PCR cleanup ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Digestion_protocol_BioBrick_Assembly_Kit Protocol#4]). | * pSB1C3 digestion and gel migration, NucleoSpin gel extract and PCR cleanup ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Digestion_protocol_BioBrick_Assembly_Kit Protocol#4]). | ||
| Line 267: | Line 53: | ||
<div id="2016-06-22" class="evday"> | <div id="2016-06-22" class="evday"> | ||
| − | + | === Wednesday, June 22th === | |
* Transformation in DH5α of ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]): | * Transformation in DH5α of ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]): | ||
| Line 280: | Line 66: | ||
<div id="2016-06-23" class="evday"> | <div id="2016-06-23" class="evday"> | ||
| − | + | === Thursday, June 23th === | |
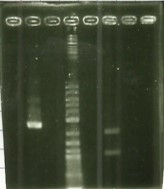
| − | + | [[File:T--Aix-Marseille--PCR-23-06.jpg|left|thumb|<u>Electrophoresis revelation</u> of E/P digestion on pSB1C3. Awaited size of 2070 bp. The two samples are the same digestion and we did a gel purification on the awaited band.]] | |
* Digestion E/P pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion E/P pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* Purification pSB1C3 purified linearized 23/06 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.236_:_Digestion_and_ligation_for_verification_and_BioBrick_Assembly Protocol#6]). | * Purification pSB1C3 purified linearized 23/06 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.236_:_Digestion_and_ligation_for_verification_and_BioBrick_Assembly Protocol#6]). | ||
| Line 289: | Line 75: | ||
* Transformation in Tg1 and DH5α ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation in Tg1 and DH5α ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| − | + | ||
</div> | </div> | ||
<div id="2016-06-24" class="evday"> | <div id="2016-06-24" class="evday"> | ||
| − | + | === Friday, June 24th === | |
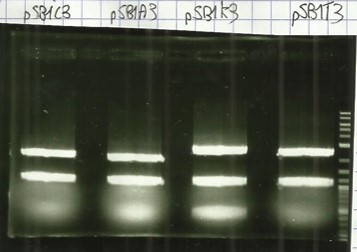
| − | + | [[File:T--Aix-Marseille--PCR-24-06.jpg|left|thumb|Verification of the plasmid purifications on pSB1C3 pSB1CA3 pSB1CK3 pSB1CT3 (the name are wroten on the top of the gel) the awaited sizes are 2070bp.]] | |
* Digestion E/P pSB1C3, pSB1A3, pSB1K3, pSB1T3, and agarose gel extract ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion E/P pSB1C3, pSB1A3, pSB1K3, pSB1T3, and agarose gel extract ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* Colony PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]) of | * Colony PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]) of | ||
| Line 305: | Line 91: | ||
* Redone transformation test device 1 and 3 interlab studies ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Redone transformation test device 1 and 3 interlab studies ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
* Plasmid purification pSB1C3 pSB1CA3 pSB1CK3 pSB1CT3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification pSB1C3 pSB1CA3 pSB1CK3 pSB1CT3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| − | |||
| − | |||
</div> | </div> | ||
<div id="2016-06-27" class="evday"> | <div id="2016-06-27" class="evday"> | ||
| − | + | === Monday, June 27th === | |
* Transformations DesA, DesB, DesC, DesD, FliC E.Coli and desulfo, interlab studies ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformations DesA, DesB, DesC, DesD, FliC E.Coli and desulfo, interlab studies ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 318: | Line 102: | ||
<div id="2016-06-28" class="evday"> | <div id="2016-06-28" class="evday"> | ||
| − | + | === Tuesday, June 28th === | |
* Plasmid purification promoter+RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification promoter+RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 326: | Line 110: | ||
<div id="2016-06-29" class="evday"> | <div id="2016-06-29" class="evday"> | ||
| − | + | === Wednesday, June 29th === | |
* Plasmid purification from interlab studies, T3 and K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification from interlab studies, T3 and K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 335: | Line 119: | ||
<div id="2016-06-30" class="evday"> | <div id="2016-06-30" class="evday"> | ||
| − | + | === Thursday, June 30th === | |
* Migration FliC coli, desulfo, DesB, DesC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Migration FliC coli, desulfo, DesB, DesC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 341: | Line 125: | ||
<div id="2016-07-01" class="evday"> | <div id="2016-07-01" class="evday"> | ||
| − | + | === Friday, July 1st === | |
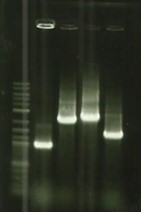
| − | + | [[File:T--Aix-Marseille--PCR-29-06.jpg|left|thumb|Verification of the plasmid purifications on pSB1C3.]] | |
* Digestion PCR products and vector pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.235_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion PCR products and vector pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.235_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* PCR clean up on digested pSB1C3, fliC coli desulfo, DesB and DesC, migration and transformation in Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | * PCR clean up on digested pSB1C3, fliC coli desulfo, DesB and DesC, migration and transformation in Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | ||
| − | |||
| − | |||
</div> | </div> | ||
<div id="2016-07-04" class="evday"> | <div id="2016-07-04" class="evday"> | ||
| − | + | === Monday, July 4th === | |
* Isolated white colonies on dishes | * Isolated white colonies on dishes | ||
| Line 356: | Line 138: | ||
<div id="2016-07-05" class="evday"> | <div id="2016-07-05" class="evday"> | ||
| − | + | === Tuesday, July 5th === | |
* Reisolated colonies from 04/07 | * Reisolated colonies from 04/07 | ||
| Line 362: | Line 144: | ||
<div id="2016-07-06" class="evday"> | <div id="2016-07-06" class="evday"> | ||
| − | + | === Wednesday, July 6th === | |
* Colony PCR with FLiC E.Coli Desulfo DesB DesC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR with FLiC E.Coli Desulfo DesB DesC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 368: | Line 150: | ||
<div id="2016-07-07" class="evday"> | <div id="2016-07-07" class="evday"> | ||
| − | + | === Thursday, July 7th === | |
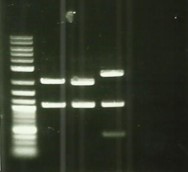
| − | + | [[File:T--Aix-Marseille--PCR-07-07.jpg|left|thumb|Colony PCR from 06/07. For the two first holes, the awaited size is 1547 bp, for the two last, this size is 994bp]] | |
* Plasmid purification DesC DesB FliC coli ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification DesC DesB FliC coli ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
* PCR on A2 A4 B1 clones from 6/7 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * PCR on A2 A4 B1 clones from 6/7 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* Digestion E/P of A5 B4 C3 C2 minipreps and pSB1A3, pSB1C3, pSB1K3, pSB1T3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion E/P of A5 B4 C3 C2 minipreps and pSB1A3, pSB1C3, pSB1K3, pSB1T3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | |||
| − | |||
</div> | </div> | ||
<div id="2016-07-08" class="evday"> | <div id="2016-07-08" class="evday"> | ||
| − | + | === Friday, July 8th === | |
* Transformation BOO34 and I0500 in pSB1K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation BOO34 and I0500 in pSB1K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 388: | Line 168: | ||
<div id="2016-07-11" class="evday"> | <div id="2016-07-11" class="evday"> | ||
| − | + | === Monday, July 11th === | |
* Redone PCR SLIC 8/8 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#.23Protocol_7_:_SLIC_.28sequence-_and_ligation-independent_cloning.29 Protocol#10]). | * Redone PCR SLIC 8/8 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#.23Protocol_7_:_SLIC_.28sequence-_and_ligation-independent_cloning.29 Protocol#10]). | ||
| Line 398: | Line 178: | ||
<div id="2016-07-12" class="evday"> | <div id="2016-07-12" class="evday"> | ||
| − | + | === Tuesday, July 12th === | |
* Purification clean up DesC and CsgA from PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | * Purification clean up DesC and CsgA from PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | ||
| Line 409: | Line 189: | ||
<div id="2016-07-13" class="evday"> | <div id="2016-07-13" class="evday"> | ||
| − | + | === Wednesday, July 13th === | |
* Plasmid purification DesA, DesB, fliC desulfo ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification DesA, DesB, fliC desulfo ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 418: | Line 198: | ||
<div id="2016-07-18" class="evday"> | <div id="2016-07-18" class="evday"> | ||
| − | + | === Monday, July 18th === | |
* Ligation (3A) in pSB1C3 of desA+RBS, desB+RBS, fliC desulfo+RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation (3A) in pSB1C3 of desA+RBS, desB+RBS, fliC desulfo+RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 429: | Line 209: | ||
<div id="2016-07-19" class="evday"> | <div id="2016-07-19" class="evday"> | ||
| − | + | === Tuesday, July 19th === | |
* Colony PCR on DesA-RBS, DesB-RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR on DesA-RBS, DesB-RBS ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 437: | Line 217: | ||
<div id="2016-07-20" class="evday"> | <div id="2016-07-20" class="evday"> | ||
| − | + | === Wednesday, July 20th === | |
* Colony PCR on DesA-RFP, DesC-RFP ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR on DesA-RFP, DesC-RFP ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 444: | Line 224: | ||
<div id="2016-07-21" class="evday"> | <div id="2016-07-21" class="evday"> | ||
| − | + | === Thursday, July 21th === | |
* Interlab studies: measurements plate reader | * Interlab studies: measurements plate reader | ||
| Line 450: | Line 230: | ||
<div id="2016-07-22" class="evday"> | <div id="2016-07-22" class="evday"> | ||
| − | + | === Friday, July 22th === | |
* Colony PCR on FliC, FliD, DesA DesB and CsgA ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR on FliC, FliD, DesA DesB and CsgA ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 456: | Line 236: | ||
<div id="2016-07-25" class="evday"> | <div id="2016-07-25" class="evday"> | ||
| − | + | === Monday, July 25th === | |
* Redone 22/7 Colony PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Redone 22/7 Colony PCR ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 464: | Line 244: | ||
<div id="2016-07-26" class="evday"> | <div id="2016-07-26" class="evday"> | ||
| − | + | === Tuesday, July 26th === | |
* Competent Tg1 preparation ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). | * Competent Tg1 preparation ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). | ||
| Line 470: | Line 250: | ||
<div id="2016-07-27" class="evday"> | <div id="2016-07-27" class="evday"> | ||
| − | + | === Wednesday, July 27th === | |
* Digestion K88005 E/S, fliC desulfo X/P, pSB1K3 E/P ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion K88005 E/S, fliC desulfo X/P, pSB1K3 E/P ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 476: | Line 256: | ||
<div id="2016-07-28" class="evday"> | <div id="2016-07-28" class="evday"> | ||
| − | + | === Thursday, July 28th === | |
* Colony PCR from respread dishes 27/7 on DesC-RBS, DesA-7.8, DesA-7.1, FliC desulfo ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR from respread dishes 27/7 on DesC-RBS, DesA-7.8, DesA-7.1, FliC desulfo ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 483: | Line 263: | ||
<div id="2016-07-29" class="evday"> | <div id="2016-07-29" class="evday"> | ||
| − | + | === Friday, July 29th === | |
* Plasmid purification DesA DesC, E/P digestion ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4] and [https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Plasmid purification DesA DesC, E/P digestion ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4] and [https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 491: | Line 271: | ||
<div id="2016-08-01" class="evday"> | <div id="2016-08-01" class="evday"> | ||
| − | + | === Monday, August 1st === | |
* Transformation ligation 29/7 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation ligation 29/7 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 498: | Line 278: | ||
<div id="2016-08-02" class="evday"> | <div id="2016-08-02" class="evday"> | ||
| − | + | === Tuesday, August 2rd === | |
| − | + | [[File:T--Aix-Marseille--PCR-02-08.jpg|left|thumb|Digestion of backbones pSB1C3, pSB1A3 and pSB1K3 verifications. Like all our backbone digestions, the awaited size is 1070bp.]] | |
* Colony PCR of CsgA from the 01/08 using EconoTac ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR of CsgA from the 01/08 using EconoTac ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* PCR clean up of DesC(200ng/µL) and gel extraction of DesA (83ng/µL), DesB(53ng/µL), FliC <i>E. coli</i> ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | * PCR clean up of DesC(200ng/µL) and gel extraction of DesA (83ng/µL), DesB(53ng/µL), FliC <i>E. coli</i> ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | ||
| − | * SLIC of DesA, DesB, DesC, DesD and transformation of 10µL in 100µL | + | * SLIC of DesA, DesB, DesC, DesD and transformation of 10µL in 100µL competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#.23Protocol_7_:_SLIC_.28sequence-_and_ligation-independent_cloning.29 Protocol#10]). |
* Sequencage of Desulfovribrio clone 3 validated. | * Sequencage of Desulfovribrio clone 3 validated. | ||
* E/P digestion on pSB1C3, pSB1A3 and pSB1K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.235_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * E/P digestion on pSB1C3, pSB1A3 and pSB1K3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.235_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | |||
| − | |||
</div><div id="2016-08-03" class="evday"> | </div><div id="2016-08-03" class="evday"> | ||
| − | + | === Wednesday, August 3th === | |
* Digestion Bba_B0034 by EcoRI/XbaI (pSB1A3) and Bba_I0500 by EcoRI/(pSB1C3) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion Bba_B0034 by EcoRI/XbaI (pSB1A3) and Bba_I0500 by EcoRI/(pSB1C3) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 518: | Line 296: | ||
</div><div id="2016-08-04" class="evday"> | </div><div id="2016-08-04" class="evday"> | ||
| − | + | === Thursday, August 4th === | |
* Transformation of Bba_K1951008 (FliC <i>E. coli</i> and spread on kanamycin LB agar petri dishes ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation of Bba_K1951008 (FliC <i>E. coli</i> and spread on kanamycin LB agar petri dishes ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
| Line 527: | Line 305: | ||
<div id="2016-08-05" class="evday"> | <div id="2016-08-05" class="evday"> | ||
| − | + | === Friday, August 5th === | |
* Colony PCR from the transformed bacteria of 08.03.2016 and starter of the good clones have been sent ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR from the transformed bacteria of 08.03.2016 and starter of the good clones have been sent ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 535: | Line 313: | ||
<div id="2016-08-08" class="evday"> | <div id="2016-08-08" class="evday"> | ||
| − | + | === Monday, August 8th === | |
| − | + | [[File:T--Aix-Marseille--PCR-08-08.jpg|left|thumb|Colony PCR verification. The clones picked up on the plates are wroten on the right of the gel. the awaited size is 506bp for all clones.]] | |
* Ligation of BBa_B0034+BBa_I0500 and Bba_K1951000 into pSB1K3 to create desA producer ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of BBa_B0034+BBa_I0500 and Bba_K1951000 into pSB1K3 to create desA producer ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | * Transformation of the previous ligation in | + | * Transformation of the previous ligation in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations protocol#2]). |
* Starters from DesC and Bba_K1951008 of the 08.04.2016. | * Starters from DesC and Bba_K1951008 of the 08.04.2016. | ||
* Resuspension of oligo using the IDT protocole. | * Resuspension of oligo using the IDT protocole. | ||
* PCR of DesB and DesC (GC enhencer, Q5 master Mix and gel extract on the good strips ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * PCR of DesB and DesC (GC enhencer, Q5 master Mix and gel extract on the good strips ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* Colony PCR of CsgA, and gel electrophoresis ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR of CsgA, and gel electrophoresis ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| − | |||
| − | |||
</div> | </div> | ||
<div id="2016-08-09" class="evday"> | <div id="2016-08-09" class="evday"> | ||
| − | + | === Tuesday, August 9th === | |
* Colony PCR from transformation of the 08.08.2016 (size waited : 3000pb), clone1 has been sent to the sequencing. | * Colony PCR from transformation of the 08.08.2016 (size waited : 3000pb), clone1 has been sent to the sequencing. | ||
| Line 558: | Line 334: | ||
<div id="2016-08-10" class="evday"> | <div id="2016-08-10" class="evday"> | ||
| − | + | === Wednesday, August 10th === | |
* Plasmid purification of Bba_K1951004 (248,49ng/µL) in pSB1K3 and sent to sequencing ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification of Bba_K1951004 (248,49ng/µL) in pSB1K3 and sent to sequencing ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 567: | Line 343: | ||
<div id="2016-08-11" class="evday"> | <div id="2016-08-11" class="evday"> | ||
| − | + | === Thursday, August 11th === | |
* Sequencing of BBa_B0034+BBa_I0500 and Bba_K1951007 have been confirmed. | * Sequencing of BBa_B0034+BBa_I0500 and Bba_K1951007 have been confirmed. | ||
| Line 576: | Line 352: | ||
<div id="2016-08-12" class="evday"> | <div id="2016-08-12" class="evday"> | ||
| − | + | === Friday, August 12th === | |
* Colony PRC from 08.10.2016 transformation again (DesB and DesD) and any clones were migrated to the good size again ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PRC from 08.10.2016 transformation again (DesB and DesD) and any clones were migrated to the good size again ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 584: | Line 360: | ||
<div id="2016-08-16" class="evday"> | <div id="2016-08-16" class="evday"> | ||
| − | + | === Tuesday, August 16th === | |
* Sequencing of Bba_K1951002 has been confirmed. | * Sequencing of Bba_K1951002 has been confirmed. | ||
| Line 593: | Line 369: | ||
<div id="2016-08-17" class="evday"> | <div id="2016-08-17" class="evday"> | ||
| − | + | === Wednesday, August 17th === | |
* Made petri dishes (10 50µG/mL kanamycin and 10 50µG/mL ampicillin). | * Made petri dishes (10 50µG/mL kanamycin and 10 50µG/mL ampicillin). | ||
| Line 602: | Line 378: | ||
<div id="2016-08-18" class="evday"> | <div id="2016-08-18" class="evday"> | ||
| − | + | === Thursday, August 18th === | |
* Plasmid purification of Bba_K1951007(203.18ng/µL), Bba_K1951003(187.53ng/µL), Bba_K1951001 (188.74ng/µL) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification of Bba_K1951007(203.18ng/µL), Bba_K1951003(187.53ng/µL), Bba_K1951001 (188.74ng/µL) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 612: | Line 388: | ||
<div id="2016-08-19" class="evday"> | <div id="2016-08-19" class="evday"> | ||
| − | + | === Friday, August 19th === | |
* Ligation of the digestion from the 08.18.2016 using 3A method in both pSB1A3 and pSB1C3 at room temperature during 1h ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of the digestion from the 08.18.2016 using 3A method in both pSB1A3 and pSB1C3 at room temperature during 1h ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 621: | Line 397: | ||
<div id="2016-08-22" class="evday"> | <div id="2016-08-22" class="evday"> | ||
| − | + | === Monday, August 22th === | |
* PCR using Q5 master with new FliC coli sequence (substitute Bbs1 site) Bba_K1951005 and Bba_K1951001, every SLIC have been validated by electrophoresis ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * PCR using Q5 master with new FliC coli sequence (substitute Bbs1 site) Bba_K1951005 and Bba_K1951001, every SLIC have been validated by electrophoresis ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* PCR clean up on the 2 previous SLIC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | * PCR clean up on the 2 previous SLIC ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | ||
| − | * SLIC and Transformation of Bba_K1951005 and Bba_K1951001 in | + | * SLIC and Transformation of Bba_K1951005 and Bba_K1951001 in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#.23Protocol_7_:_SLIC_.28sequence-_and_ligation-independent_cloning.29 Protocol#10]). |
* Starter of Bba_K1951004, Bba_K1951009 and Bba_K1951010 to make a protein production test. | * Starter of Bba_K1951004, Bba_K1951009 and Bba_K1951010 to make a protein production test. | ||
</div> | </div> | ||
<div id="2016-08-23" class="evday"> | <div id="2016-08-23" class="evday"> | ||
| − | + | === Tuesday, August 23th === | |
* Colony PCR from transformation of the 08.25.2016 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR from transformation of the 08.25.2016 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 640: | Line 416: | ||
<div id="2016-08-24" class="evday"> | <div id="2016-08-24" class="evday"> | ||
| − | + | === Wednesday, August 24th === | |
* Plasmid purification from Bba_K1951008, Bba_K1951009 and Bba_K1951001 and send to the sequencing with oligo FW ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification from Bba_K1951008, Bba_K1951009 and Bba_K1951001 and send to the sequencing with oligo FW ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 647: | Line 423: | ||
<div id="2016-08-25" class="evday"> | <div id="2016-08-25" class="evday"> | ||
| − | + | === Thursday, August 25th === | |
* Digestion of Bba_K1951002 and Bba_K1951003 by XbaI and PstI, Bba_B0034 by SpeI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion of Bba_K1951002 and Bba_K1951003 by XbaI and PstI, Bba_B0034 by SpeI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 653: | Line 429: | ||
<div id="2016-08-25" class="evday"> | <div id="2016-08-25" class="evday"> | ||
| − | + | === Friday, August 26th === | |
* Sequencing Bba_K1951009, Bba_K1951001, Bba_K1951008 and Bba_K1951010 ok with the FW oligo only. | * Sequencing Bba_K1951009, Bba_K1951001, Bba_K1951008 and Bba_K1951010 ok with the FW oligo only. | ||
* Preparation of the HPLC solution : | * Preparation of the HPLC solution : | ||
| Line 676: | Line 452: | ||
* Pcr clean up on the digestion from the 08.25.2016 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | * Pcr clean up on the digestion from the 08.25.2016 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.237_:_PCR_clean-up_and_gel_extraction Protocol#7]). | ||
* Ligation using 2A method of Bba_K1951002 or Bba_K1951003 and Bba_B0034 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation using 2A method of Bba_K1951002 or Bba_K1951003 and Bba_B0034 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | * Transformation of the previous ligation in | + | * Transformation of the previous ligation in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Digestion of 500ng in 50µL total mix of Bba_K1951002 and Bba_K1951005 by XbaI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion of 500ng in 50µL total mix of Bba_K1951002 and Bba_K1951005 by XbaI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* Ligation of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) overnight at 16°C ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) overnight at 16°C ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| Line 683: | Line 459: | ||
<div id="2016-08-29" class="evday"> | <div id="2016-08-29" class="evday"> | ||
| − | + | === Monday, August 29th === | |
* Sequencing Bba_K1951001, Bba_K1951005, Bba_K1951009 and Bba_K1951010 ok with the RV oligo only so VALIDATE. | * Sequencing Bba_K1951001, Bba_K1951005, Bba_K1951009 and Bba_K1951010 ok with the RV oligo only so VALIDATE. | ||
| Line 692: | Line 468: | ||
<div id="2016-08-30" class="evday"> | <div id="2016-08-30" class="evday"> | ||
| − | + | === Tuesday, August 30th === | |
| − | + | [[File:T--Aix-Marseille--PCR-30-06.jpg|left|thumb|Migration of FliC from E.coli, FliC from desulfo, DesB and DesC. The awaited size for the 8 DesD+RBS is 2150bp, for the FliC 25-54 it's 1900bp.]] | |
* Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) + BBa_I0500 (Prom) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) + BBa_I0500 (Prom) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| − | * Transformation of Bba_K1951004 (DesA with pSB1C3) in | + | * Transformation of Bba_K1951004 (DesA with pSB1C3) in competent Tg1 cells ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Ligation of Bba_K1951004 digested E/P in pSB1C3 at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of Bba_K1951004 digested E/P in pSB1C3 at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
NB : we wanted to transform a cadA mutant from KEIO bank so we can't use this biobrick in pSB1K3. | NB : we wanted to transform a cadA mutant from KEIO bank so we can't use this biobrick in pSB1K3. | ||
* Starters from intermediate biobirck Bba_K1951001 (DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) and Bba_K1951000(DesA),Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom). | * Starters from intermediate biobirck Bba_K1951001 (DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) and Bba_K1951000(DesA),Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom). | ||
| − | * Made | + | * Made competent cells Tg1, culture from an over night starter have grown up from Abs(600nm)=0.05 until Abs(600nm)=0.457 at 37°C ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). |
| − | + | ||
| − | + | ||
</div> | </div> | ||
<div id="2016-09-01" class="evday"> | <div id="2016-09-01" class="evday"> | ||
| − | + | === Thursday, September 1st === | |
* Plasmid purification of the starter Bba_K1951001(175ng/µL), Bba_K1951002(250ng/µL), Bba_K1951003(440ng/µL) and Bba_K1951005(183ng/µL) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | * Plasmid purification of the starter Bba_K1951001(175ng/µL), Bba_K1951002(250ng/µL), Bba_K1951003(440ng/µL) and Bba_K1951005(183ng/µL) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Plasmid_DNA_purification Protocol#4]). | ||
| Line 713: | Line 487: | ||
* Ligation of Bba_K1951004 (S/P) and Bba_K1951001 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of Bba_K1951004 (S/P) and Bba_K1951001 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* Ligation of Bba_K1951002 + RBS (E/S) and Bba_K1951003 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of Bba_K1951002 + RBS (E/S) and Bba_K1951003 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | * Made | + | * Made competent cells of fliC mutant and cadA mutant from the Keio bank and test on different antibiotic petri dishes (only kanamycin dish was a positive control) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). |
* Retest of colony PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) and nothing was good ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Retest of colony PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) and nothing was good ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| − | * Transformation of the ligation Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) from the ligation of the 08.26.2016 with the new | + | * Transformation of the ligation Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) from the ligation of the 08.26.2016 with the new competent cells from 08.30.2016 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
| − | * Transformation of Bba_K1951008 in fliC mutant Keio | + | * Transformation of Bba_K1951008 in fliC mutant Keio competent made the 30th, spread on Kana/Cm petri dishes ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Made a calibration range for HPLC. | * Made a calibration range for HPLC. | ||
</div> | </div> | ||
<div id="2016-09-02" class="evday"> | <div id="2016-09-02" class="evday"> | ||
| − | + | === Friday, September 2rd === | |
* Remade ligation of pSB1C3 (E/P) and Bba_K1951004 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Remade ligation of pSB1C3 (E/P) and Bba_K1951004 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | * Transformation of the previous ligation mix in | + | * Transformation of the previous ligation mix in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Colony PCR on transformation of intermediate biobricks ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]).: | * Colony PCR on transformation of intermediate biobricks ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]).: | ||
- Bba_K1951004 + RBS (X/P) Bba_K1951001 in pSB1K3(E/P) at 16°C overnight | - Bba_K1951004 + RBS (X/P) Bba_K1951001 in pSB1K3(E/P) at 16°C overnight | ||
| Line 731: | Line 505: | ||
<div id="2016-09-05" class="evday"> | <div id="2016-09-05" class="evday"> | ||
| − | + | === Monday, September 5th === | |
* Analysis of the electrophoresis results from 09.02.2016. | * Analysis of the electrophoresis results from 09.02.2016. | ||
| Line 741: | Line 515: | ||
<div id="2016-09-06" class="evday"> | <div id="2016-09-06" class="evday"> | ||
| − | + | === Tuesday, September 6th === | |
| − | + | [[File:T--Aix-Marseille--PCR-06-09.jpg|left|thumb|PCR colony verification. The holes 2 to 7 are the clone 12 of the plate and 11 to 16 are the clone 13 of the plate. The holes 9 and 17 are the RPF (control).]] | |
| − | * Transformation of cadA mutant | + | * Transformation of cadA mutant competent strain by Bba_K1951004 in pSB1C3, spread on Kana/Cm petri dishes ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Colony PCR Bba_K1951004 in pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR Bba_K1951004 in pSB1C3 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
* Starters from Bba_K1951004 in pSB1C3. | * Starters from Bba_K1951004 in pSB1C3. | ||
* 1 step of the transduction using phage p1 from fliC mutant Keio bank. Conserved at 4°C with 20µL chlorophorme | * 1 step of the transduction using phage p1 from fliC mutant Keio bank. Conserved at 4°C with 20µL chlorophorme | ||
* PCR with Q5 master mix using oligo of diriged mutagenesis for the insertion of Bbs1 site in the 214-215 and 238-239 sites of BBa_K1951008. Digest 3h with addition of 1µL DpNI in the PCR tube ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * PCR with Q5 master mix using oligo of diriged mutagenesis for the insertion of Bbs1 site in the 214-215 and 238-239 sites of BBa_K1951008. Digest 3h with addition of 1µL DpNI in the PCR tube ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| − | * Transformation of the previous PCR in | + | * Transformation of the previous PCR in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Starters of the PCR from 09.02.2016. | * Starters of the PCR from 09.02.2016. | ||
* Remade PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) but still no result ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Remade PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) but still no result ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| − | |||
| − | |||
</div> | </div> | ||
<div id="2016-09-07" class="evday"> | <div id="2016-09-07" class="evday"> | ||
| − | + | === Wednesday, September 7th === | |
* Colony PCR on cadA mutant complemented with BBa_K1951004 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | * Colony PCR on cadA mutant complemented with BBa_K1951004 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_PCR_and_electrophoresis Protocol#3]). | ||
| Line 765: | Line 537: | ||
<div id="2016-09-08" class="evday"> | <div id="2016-09-08" class="evday"> | ||
| − | + | === Thursday, September 8th === | |
* Starters of BBa_K1951008 + BbsI 215-216 position and starters of BBa_K1951008 + BbsI 238-239 position. | * Starters of BBa_K1951008 + BbsI 215-216 position and starters of BBa_K1951008 + BbsI 238-239 position. | ||
| Line 771: | Line 543: | ||
* Digestion of intermediate biobrick BBa_K1951004+ RBS BBa_K1951001 by EcoRI and SpeI and RBS BBa_K1951001+ RBS BBa_K1951002 by SpeI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Digestion of intermediate biobrick BBa_K1951004+ RBS BBa_K1951001 by EcoRI and SpeI and RBS BBa_K1951001+ RBS BBa_K1951002 by SpeI and PstI ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
* Ligation of the 2 previous digestions in pSB1C3 to make the final BBa_K1951011 (E/P) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | * Ligation of the 2 previous digestions in pSB1C3 to make the final BBa_K1951011 (E/P) ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.233_:_Cloning_protocol_for_IDT_sequences Protocol#5]). | ||
| − | * Transformation of the previous ligation in | + | * Transformation of the previous ligation in competent Tg1 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
</div> | </div> | ||
<div id="2016-09-09" class="evday"> | <div id="2016-09-09" class="evday"> | ||
| − | + | === Friday, September 9th === | |
* Derivatization of the sample cadA mutant complemented with BBa_K1951004 and cadA mutant complemented with BBa_K1951004+ RBS BBa_K1951001 . | * Derivatization of the sample cadA mutant complemented with BBa_K1951004 and cadA mutant complemented with BBa_K1951004+ RBS BBa_K1951001 . | ||
| Line 785: | Line 557: | ||
<div id="2016-09-10" class="evday"> | <div id="2016-09-10" class="evday"> | ||
| − | + | === Satursday, September 10th === | |
* Swimming test of the knockout fliC mutant W3110 for 10 differents colonies and the W3110 WT as a positive temoin ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.2311_:_Swimming_test Protocol#11]). | * Swimming test of the knockout fliC mutant W3110 for 10 differents colonies and the W3110 WT as a positive temoin ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.2311_:_Swimming_test Protocol#11]). | ||
| Line 792: | Line 564: | ||
<div id="2016-09-11" class="evday"> | <div id="2016-09-11" class="evday"> | ||
| − | + | === Sunday, September 11st === | |
| − | * Made | + | * Made competent of the fliC mutant W3110 strain from 5 differents colonies from 4 colonies which swam well ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.231_:_Preparation_of_competent_bacteria_cells Protocol#1]). |
| − | * Transformation of the previous | + | * Transformation of the previous competent with BBa_K1951008 and BBa_K1951009 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
* Transformation of cadA mutant Keio strain with BBa_K1951011 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | * Transformation of cadA mutant Keio strain with BBa_K1951011 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). | ||
</div> | </div> | ||
<div id="2016-09-12" class="evday"> | <div id="2016-09-12" class="evday"> | ||
| − | + | === Monday, September 12rd === | |
| − | * Transformation of the fliC mutant W3110 | + | * Transformation of the fliC mutant W3110 competent with BBa_K1951008 ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.232_:_Transformations Protocol#2]). |
</div> | </div> | ||
<div id="2016-09-13" class="evday"> | <div id="2016-09-13" class="evday"> | ||
| − | + | === Tuesday, September 13th === | |
* Swimming test of colonies obtained after transformation of the 09.12.2016. After 3h incubation at 37°C, swimming has been recovered ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.2311_:_Swimming_test Protocol#11]). | * Swimming test of colonies obtained after transformation of the 09.12.2016. After 3h incubation at 37°C, swimming has been recovered ([https://2016.igem.org/Team:Aix-Marseille/Experiments/Protocols#Protocol_.2311_:_Swimming_test Protocol#11]). | ||
| Line 814: | Line 586: | ||
<div id="2016-09-15" class="evday"> | <div id="2016-09-15" class="evday"> | ||
| − | + | === Thursday, September 15=== | |
* Dillution of peptides oligos by 10000 (ex: 0.31mg/238µL/10000=ng/µL). | * Dillution of peptides oligos by 10000 (ex: 0.31mg/238µL/10000=ng/µL). | ||
Monday, June 13th
- Preparation of heat competent DH5α cells (Protocol #1).
Tuesday, June 14th
- Transformation of DH5α cells with test plasmid (Protocol#2).
Wednesday, June 15th
- Preparation of heat competent Tg1 cells (Protocol#1).
- Transformation of Tg1 cells with test plasmid (Protocol#2).
Thursday, June 16th
- Transformation of DH5α cells with pSB1C3 + RFP (Protocol#2).
- Transformation with K880005, K823017, I0500, K1404006, B0034 (Protocol#2).
Friday, June 17th
- Plasmids purification K880005, K823017, I0500, K1404006, B0034 (Protocol#4).
- Plasmids purification pSB1C3 + RFP (Protocol#4).
Monday, June 20th
- pSB1C3 digestion from 17/06 plasmids purification (Protocol#4).
- pSB1C3 digestion gel migration (Protocol#6 and Protocol#3).
- Biobricks plasmids purifications gel migration (Protocol#4).
- Transformation Tg1 with RFP+pSB1C3 (Protocol#2)
Tuesday, June 21th
- pSB1C3 digestion and gel migration, NucleoSpin gel extract and PCR cleanup (Protocol#4).
- E. Coli and Desulfo FliC (E/P digested) ligation in linearized pSB1C3 (Protocol#5).
- Transformation of pSB1C3 RFP in Tg1 (Protocol#2).
Wednesday, June 22th
- Transformation in DH5α of (Protocol#2):
- FliC coli/desulfo in pSB1C3
- Linearized pSB1K3
- Linearized pSB1A3
- Linearized pSB1T3
- Linearized pSB1C3
- Plasmid purification B0034 (Protocol#4).
- Transformation in DH5α of interlab studies plasmids (Protocol#2).
Thursday, June 23th
- Digestion E/P pSB1C3 (Protocol#5).
- Purification pSB1C3 purified linearized 23/06 (Protocol#6).
- Digestion E/P of DesA, DesB, DesC, DesD (Protocol#5).
- 3A Assembly of B0034 and I0500 in pSB1K3 (Protocol#6).
- Digestion with pSB1C3 23/06 insert digested fliC (Protocol#5).
- Transformation in Tg1 and DH5α (Protocol#2).
Friday, June 24th
- Digestion E/P pSB1C3, pSB1A3, pSB1K3, pSB1T3, and agarose gel extract (Protocol#5).
- Colony PCR (Protocol#3) of
- RBS+promoter
- GFP
- empty
- Ligation: FliC E.Coli, FliC desulfo, DesA B C D (Protocol#3).
- Ligation pSB1C3 and fliC E.Coli / desulfo digested E/P 21/06 (Protocol#3).
- Transformation in Tg1 and DH5α (Protocol#2).
- Redone transformation test device 1 and 3 interlab studies (Protocol#2).
- Plasmid purification pSB1C3 pSB1CA3 pSB1CK3 pSB1CT3 (Protocol#4).
Monday, June 27th
- Transformations DesA, DesB, DesC, DesD, FliC E.Coli and desulfo, interlab studies (Protocol#2).
- Transformations failed (Protocol#2).
- Transformation pSB1T3 plasmid purification, pSB1K3 plasmid purification, control (Protocol#4 andProtocol#2).
Tuesday, June 28th
- Plasmid purification promoter+RBS (Protocol#4).
- PCR of IDT sequences (fliC and DesA to D) and gel check (Protocol#3).
- Gel extract pSB1CK3, pSB1CC3, pSB1CT3 (Protocol#7).
Wednesday, June 29th
- Plasmid purification from interlab studies, T3 and K3 (Protocol#4).
- Ligation and transformation in Tg1 Sandra Des A to D, fliC coli desulfo (Protocol#3 and Protocol#2).
- Redone PCR of IDT sequences (fliC and DesA to D) (different Tm) and gel extract (Protocol#3 and Protocol#7).
- Transformation of fliCs from remaining ligation in Tg1 Sandra (Protocol#2).
Thursday, June 30th
- Migration FliC coli, desulfo, DesB, DesC (Protocol#3).
Friday, July 1st
- Digestion PCR products and vector pSB1C3 (Protocol#5).
- PCR clean up on digested pSB1C3, fliC coli desulfo, DesB and DesC, migration and transformation in Tg1 (Protocol#7).
Monday, July 4th
- Isolated white colonies on dishes
Tuesday, July 5th
- Reisolated colonies from 04/07
Wednesday, July 6th
- Colony PCR with FLiC E.Coli Desulfo DesB DesC (Protocol#3).
Thursday, July 7th
- Plasmid purification DesC DesB FliC coli (Protocol#4).
- PCR on A2 A4 B1 clones from 6/7 (Protocol#3).
- Digestion E/P of A5 B4 C3 C2 minipreps and pSB1A3, pSB1C3, pSB1K3, pSB1T3 (Protocol#5).
Friday, July 8th
- Transformation BOO34 and I0500 in pSB1K3 (Protocol#2).
- Gel verification of 7/7 digestion (Protocol#6).
- Plasmid purification A5 (fliC coli) and C2 (DesB) from 7/7 starters (Protocol#4).
- PCR clean up of vectors (Protocol#7).
- PCR SLIC FliC coli/desulfo, DesA, B, C, D, csgA -> failed (Protocol#3).
Monday, July 11th
- Redone PCR SLIC 8/8 (Protocol#10).
- Redone PCR DesC, DesD and csgA (Protocol#3).
- SLIC desA, desB, fliC coli desulfo in pSB1A3 (Protocol#10).
- Transformation Tg1 with fliC coli desulfo, desA, desB in pSB1A3 from SLIC (Protocol#2).
- PCR clean up desA, desB, fliC coli desulfo (Protocol#7).
Tuesday, July 12th
- Purification clean up DesC and CsgA from PCR (Protocol#7).
- Digestion E/P pSB1A3, pSB1K3, pSB1C3, PCR clean up (Protocol#5).
- Colony PCR SLIC 11/07 FliC desulfo, desA, desB (Protocol#10).
- SLIC in pSB1C3 of DesA/B/C, FliC coli and desulfo, csgA (Protocol#10).
- Transformation SLIC mix in Tg1 (Protocol#2).
- Transformation constructions #7 (kanamycin) 9 and 55 (chloramphenicol) (Protocol#2).
Wednesday, July 13th
- Plasmid purification DesA, DesB, fliC desulfo (Protocol#4).
- Digestion and PCR clean up of pSB1C3 E/P, B0034 E/S, K88005 S/P, fliC desulfo X/P, desA X/P, desB X/P (Protocol#5).
- Colony PCR #1 2 3 50 51 53 (Protocol#3).
- Plasmid purification pSB1 A/C/K/T 3, gel migration (Protocol#4).
Monday, July 18th
- Ligation (3A) in pSB1C3 of desA+RBS, desB+RBS, fliC desulfo+RBS (Protocol#5).
- Transformation in Tg1 (Protocol#2).
- Plasmid purification empty vectors C3 A3 K3 (Protocol#4).
- Interlab studies : resuspended FITC, incubation 42°C.
- Digestion E/P and PCR clean up empty vectors A3 C3 K3 (Protocol#5).
- PCR clean up on A3, C3 and K3 (Protocol#7).
Tuesday, July 19th
- Colony PCR on DesA-RBS, DesB-RBS (Protocol#3).
- Ligation and transformation in Tg1 (Protocol#5 and Protocol#2).
- Interlab studies: transformation in DH5α (Protocol#2).
Wednesday, July 20th
- Colony PCR on DesA-RFP, DesC-RFP (Protocol#3).
- Plasmid purification gel migration DesB rbs, DesA rbs, DesB (Protocol#4).
Thursday, July 21th
- Interlab studies: measurements plate reader
Friday, July 22th
- Colony PCR on FliC, FliD, DesA DesB and CsgA (Protocol#3).
Monday, July 25th
- Redone 22/7 Colony PCR (Protocol#3).
- Plasmid purification DesC Csg1 DesC+RBS and digestion E/P (Protocol#4).
- Digestion E/P DesA+RBS, DesB (Protocol#5).
Tuesday, July 26th
- Competent Tg1 preparation (Protocol#1).
Wednesday, July 27th
- Digestion K88005 E/S, fliC desulfo X/P, pSB1K3 E/P (Protocol#5).
Thursday, July 28th
- Colony PCR from respread dishes 27/7 on DesC-RBS, DesA-7.8, DesA-7.1, FliC desulfo (Protocol#3).
- PCR gradient on csgA (BBa140034) (Protocol#3).
Friday, July 29th
- Plasmid purification DesA DesC, E/P digestion (Protocol#4 and Protocol#5).
- Ligation 3A pSB1K3 RBS FliC desulfo (Protocol#5).
- Redone gel migration PCR 28/7 (Protocol#3).
Monday, August 1st
- Transformation ligation 29/7 (Protocol#2).
- PCR amplification IDT sequences DesA/B/C fliC coli and oligo, gel extract (Protocol#3 and Protocol#7).
Tuesday, August 2rd
- Colony PCR of CsgA from the 01/08 using EconoTac (Protocol#3).
- PCR clean up of DesC(200ng/µL) and gel extraction of DesA (83ng/µL), DesB(53ng/µL), FliC E. coli (Protocol#7).
- SLIC of DesA, DesB, DesC, DesD and transformation of 10µL in 100µL competent Tg1 (Protocol#10).
- Sequencage of Desulfovribrio clone 3 validated.
- E/P digestion on pSB1C3, pSB1A3 and pSB1K3 (Protocol#5).
Wednesday, August 3th
- Digestion Bba_B0034 by EcoRI/XbaI (pSB1A3) and Bba_I0500 by EcoRI/(pSB1C3) (Protocol#5).
- Ligation overnight at 16°C (Protocol#5).
- by 2A technical : BBa_B0034+BBa_I0500 - by 3A technical : Bba_K1951005 and Bba_K880005 digested the 07.18.2016
- Transformation of BBa_B0034+BBa_I0500 ligated in Tg1 and stread on Amp LB agar petri dishes (Protocol#2).
Thursday, August 4th
- Transformation of Bba_K1951008 (FliC E. coli and spread on kanamycin LB agar petri dishes (Protocol#2).
- From SLIC of the 08.02.2016 and 2 ligations, colonies of DesB and DesC have been repatched on chloramphenicol LB agar petri dishes.
- Colony PCR from the transformed bacteria of 08.03.2016 and starter of the good clones have been sent (Protocol#3).
- Plasmid purification of DesA, clone3(141.47ng/µL), FliC E. coli clone 2 (169,76ng/µL) and FliC Desulfo (133,47ng/µL) valided to the sequencage have been made (Protocol#4).
Friday, August 5th
- Colony PCR from the transformed bacteria of 08.03.2016 and starter of the good clones have been sent (Protocol#3).
- SLIC of CsgA in pSB1C3 (Protocol#10).
- Sent to the sequencing Bba_K1951009 and BBa_B0034+BBa_I0500.
Monday, August 8th
- Ligation of BBa_B0034+BBa_I0500 and Bba_K1951000 into pSB1K3 to create desA producer (Protocol#5).
- Transformation of the previous ligation in competent Tg1 (protocol#2).
- Starters from DesC and Bba_K1951008 of the 08.04.2016.
- Resuspension of oligo using the IDT protocole.
- PCR of DesB and DesC (GC enhencer, Q5 master Mix and gel extract on the good strips (Protocol#3).
- Colony PCR of CsgA, and gel electrophoresis (Protocol#3).
Tuesday, August 9th
- Colony PCR from transformation of the 08.08.2016 (size waited : 3000pb), clone1 has been sent to the sequencing.
- Plasmid purification of Bba_K1951008 (126.52ng/µL), Bba_K1951002 (102.18ng/µL), Bba_K1951007 (8,33ng/µL) (Protocol#4).
- From the IDT sequence and gel extract of the 08.08.2016, PCR using Q5 master Mix with 2 different programs on DesB and DesD
- 98°C 30s, (98°C 10s, 60°C 30s, 72°C 1min)*30, 72°C 2min. - 98°C 30s, (98°C 10s, 72°C 1min)*30, 72°C 2min.
Wednesday, August 10th
- Plasmid purification of Bba_K1951004 (248,49ng/µL) in pSB1K3 and sent to sequencing (Protocol#4).
- Glyceroled tubes of Bba_K1951004 (248,49ng/µL) in pSB1K3.
- PCR clean up realised on PCR from the 08.09.2016, DesB (35ng/µL) and DesD (48ng/µL) (Protocol#7).
- SLIC realised from the previous PCR in pSB1C3 and transformation in Tg1 (Protocol#10).
Thursday, August 11th
- Sequencing of BBa_B0034+BBa_I0500 and Bba_K1951007 have been confirmed.
- Colony PRC from 08.10.2016 transformation and any clones were migrated to the good size (Protocol#3).
- Digestion of Bba_K1951007 by PstI and XbaI at 37°C during 1h (Protocol#5).
- Ligation over night of Bba_K1951007 and Bba_K880005 to create Bba_K1951010 (CsgA producer) (Protocol#5).
Friday, August 12th
- Colony PRC from 08.10.2016 transformation again (DesB and DesD) and any clones were migrated to the good size again (Protocol#3).
- Plasmid purification on Bba_K1951009 (116.84ng/µL) (Protocol#4).
- Ligation over night of Bba_K1951007 and Bba_K880005 to create Bba_K1951010 (CsgA producer) (Protocol#4).
Tuesday, August 16th
- Sequencing of Bba_K1951002 has been confirmed.
- Plasmid purification on Bba_K1951009 (116.84ng/µL) (Protocol#4).
- Colony PCR of Bba_K1951001 and Bba_K1951003 (Protocol#3).
- Screening on agarose gel; 7 clones have been mixed in a PCR tube, and mix in 1µL used as DNA template.
Wednesday, August 17th
- Made petri dishes (10 50µG/mL kanamycin and 10 50µG/mL ampicillin).
- Colony PCR of Bba_K1951007 (Protocol#3).
- SLIC of DesB and DesD in pSB1C3 (Protocol#10).
- Transformation of the previous SLIC in compétante Tg1 (Protocol#2).
Thursday, August 18th
- Plasmid purification of Bba_K1951007(203.18ng/µL), Bba_K1951003(187.53ng/µL), Bba_K1951001 (188.74ng/µL) (Protocol#4).
- Glyceroled tubes on the previous biobricks.
- Previous biobrick sent to the sequencing.
- Digestion of Bba_K1951005, Bba_K1951006, Bba_K1951005 by XbaI and PstI ; Bba_K1951007 by SpeI and PstI (Protocol#5).
- Migration of the previous biobrick (size waited : Bba_K1951005=1547pb, Bba_K1951006=944pb, Bba_K1951007=506pb).
Friday, August 19th
- Ligation of the digestion from the 08.18.2016 using 3A method in both pSB1A3 and pSB1C3 at room temperature during 1h (Protocol#5).
- Transformation of the previous ligation in Tg1 (Protocol#2).
- Colony PCR on the SLIC transformation of the 08.17.2016, nothing has worked (Protocol#3).
- Sequencing of the Bba_K1951004 is ok (desA producer).
Monday, August 22th
- PCR using Q5 master with new FliC coli sequence (substitute Bbs1 site) Bba_K1951005 and Bba_K1951001, every SLIC have been validated by electrophoresis (Protocol#3).
- PCR clean up on the 2 previous SLIC (Protocol#7).
- SLIC and Transformation of Bba_K1951005 and Bba_K1951001 in competent Tg1 (Protocol#10).
- Starter of Bba_K1951004, Bba_K1951009 and Bba_K1951010 to make a protein production test.
Tuesday, August 23th
- Colony PCR from transformation of the 08.25.2016 (Protocol#3).
- From starter of the 08.22.2016, culture was started at A(600nm) = 0.1 and induced with 0.02% arabinose at A(600nm)=0.5. After 2h incubation, DO was taken again and 1uDO was sampled. The sample was centrifiged at 5000g during 5min. Supernant was removed and pellet resuspended in 50µL of charge buffer. 5µL was charged on a polyacrylamide gel.
- Plasmid purification from Bba_K1951003 (171ng/µL), Bba_K1951004(217ng/µL), Bba_K1951007(139ng/µL) (Protocol#4).
- Colony PCR from Bba_K1951008, Bba_K1951009
- Starters from Bba_K1951008 and Bba_K1951009 that seem ok.
Wednesday, August 24th
- Plasmid purification from Bba_K1951008, Bba_K1951009 and Bba_K1951001 and send to the sequencing with oligo FW (Protocol#4).
- Colony PCR from Bba_K1951005, Bba_K1951008, Bba_K1951009 and Bba_K1951010 (Protocol#3).
Thursday, August 25th
- Digestion of Bba_K1951002 and Bba_K1951003 by XbaI and PstI, Bba_B0034 by SpeI and PstI (Protocol#5).
Friday, August 26th
- Sequencing Bba_K1951009, Bba_K1951001, Bba_K1951008 and Bba_K1951010 ok with the FW oligo only.
- Preparation of the HPLC solution :
- Buffer Borate 0.05M pH9 ( ajusted with NaOH)
- L-Lysine 0.01M
- DEEMM 0.2M
- Hexamethylenediamine 0.01M
- Test of the HPLC C18 column with 3 mixs :
1 (Lysine): Borate 180µL, Methanol 60µL, H2O ppi 47µL, Lysine 30µL, DEEMM 3µL
2 (Hexamethylenediamine): Borate 180µL, Methanol 60µL, H2O ppi 47µL, Hexamethylenediamine 30µL, DEEMM 3µL
3 (Negative temoin): Borate 180µL, Methanol 60µL, H2O ppi 77µL, DEEMM 3µL
Heat to 70°C during 2 hours
- Pcr clean up on the digestion from the 08.25.2016 (Protocol#7).
- Ligation using 2A method of Bba_K1951002 or Bba_K1951003 and Bba_B0034 (Protocol#5).
- Transformation of the previous ligation in competent Tg1 (Protocol#2).
- Digestion of 500ng in 50µL total mix of Bba_K1951002 and Bba_K1951005 by XbaI and PstI (Protocol#5).
- Ligation of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) overnight at 16°C (Protocol#5).
- Ligation of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) + BBa_I0500 (Prom) overnight at 16°C (Protocol#5).
Monday, August 29th
- Sequencing Bba_K1951001, Bba_K1951005, Bba_K1951009 and Bba_K1951010 ok with the RV oligo only so VALIDATE.
- Digestion of 500ng in 50µL total mix of Bba_K1951004 EcoRI and PstI (Protocol#5).
- Ligation of Bba_K1951004 digested E/P in pSB1C3 at 16°C overnight (Protocol#5).
NB : we wanted to transform a cadA mutant from KEIO bank so we can't use this biobrick in pSB1K3.
Tuesday, August 30th
- Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) (Protocol#3).
- Colony PCR of Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) and Bba_B0034 (rbs) + BBa_I0500 (Prom) (Protocol#3).
- Transformation of Bba_K1951004 (DesA with pSB1C3) in competent Tg1 cells (Protocol#2).
- Ligation of Bba_K1951004 digested E/P in pSB1C3 at 16°C overnight (Protocol#5).
NB : we wanted to transform a cadA mutant from KEIO bank so we can't use this biobrick in pSB1K3.
- Starters from intermediate biobirck Bba_K1951001 (DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) and Bba_K1951000(DesA),Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom).
- Made competent cells Tg1, culture from an over night starter have grown up from Abs(600nm)=0.05 until Abs(600nm)=0.457 at 37°C (Protocol#1).
Thursday, September 1st
- Plasmid purification of the starter Bba_K1951001(175ng/µL), Bba_K1951002(250ng/µL), Bba_K1951003(440ng/µL) and Bba_K1951005(183ng/µL) (Protocol#4).
- Glyceroled tubes from starter Bba_K1951001(175ng/µL), Bba_K1951002(250ng/µL), Bba_K1951003(440ng/µL) and Bba_K1951005(183ng/µL)and from starter of cadA mutant strain and fliC mutant strain from Keio bank.
- Digestion of intermediate biobricks BBa_K1951004 by SpeI/PstI ; Bba_K1951001 (DesB) and Bba_K1951003(DesD)with BBa_B0034 (RBS) by XbaI, PstI ; Bba_K1951002 (DesC) with Bba_B0034 (RBS) by EcoRI/SpeI (Protocol#5).
- Ligation of Bba_K1951004 (S/P) and Bba_K1951001 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight (Protocol#5).
- Ligation of Bba_K1951002 + RBS (E/S) and Bba_K1951003 + RBS (X/P) in pSB1K3(E/P) at 16°C overnight (Protocol#5).
- Made competent cells of fliC mutant and cadA mutant from the Keio bank and test on different antibiotic petri dishes (only kanamycin dish was a positive control) (Protocol#1).
- Retest of colony PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) and nothing was good (Protocol#3).
- Transformation of the ligation Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) from the ligation of the 08.26.2016 with the new competent cells from 08.30.2016 (Protocol#2).
- Transformation of Bba_K1951008 in fliC mutant Keio competent made the 30th, spread on Kana/Cm petri dishes (Protocol#2).
- Made a calibration range for HPLC.
Friday, September 2rd
- Remade ligation of pSB1C3 (E/P) and Bba_K1951004 (Protocol#5).
- Transformation of the previous ligation mix in competent Tg1 (Protocol#2).
- Colony PCR on transformation of intermediate biobricks (Protocol#3).:
- Bba_K1951004 + RBS (X/P) Bba_K1951001 in pSB1K3(E/P) at 16°C overnight - RBS Bba_K1951002(E/S) and RBS Bba_K1951003(X/P) in pSB1K3(E/P) at 16°C overnight
Monday, September 5th
- Analysis of the electrophoresis results from 09.02.2016.
- Swimming test of the WT Keio strain and the fliC mutant keio strain. Nor the WT neither the fliC mutant strain are abled to swim. We decide to make a fliC mutant from a good swimmer strain (Escherichia coli W3110) (Protocol#11).
- Colony PCR Bba_K1951004 in pSB1C3 (Protocol#3)
- Starters from Bba_K1951004 in pSB1C3.
- 1 step of the transduction using phage p1 from fliC mutant Keio bank. Conserved at 4°C with 20µL chlorophorme.
Tuesday, September 6th
- Transformation of cadA mutant competent strain by Bba_K1951004 in pSB1C3, spread on Kana/Cm petri dishes (Protocol#2).
- Colony PCR Bba_K1951004 in pSB1C3 (Protocol#3).
- Starters from Bba_K1951004 in pSB1C3.
- 1 step of the transduction using phage p1 from fliC mutant Keio bank. Conserved at 4°C with 20µL chlorophorme
- PCR with Q5 master mix using oligo of diriged mutagenesis for the insertion of Bbs1 site in the 214-215 and 238-239 sites of BBa_K1951008. Digest 3h with addition of 1µL DpNI in the PCR tube (Protocol#3).
- Transformation of the previous PCR in competent Tg1 (Protocol#2).
- Starters of the PCR from 09.02.2016.
- Remade PCR on Bba_K1951001(DesB), Bba_K1951002(DesC), Bba_K1951003(DesD) with Bba_B0034 (rbs) + BBa_I0500(Prom) but still no result (Protocol#3).
Wednesday, September 7th
- Colony PCR on cadA mutant complemented with BBa_K1951004 (Protocol#3).
- HPLC test of cadA mutant complemented with BBa_K1951004 (Protocol#9).
- Colony PCR to verify the insertion of Bbs1 site in the 214-215 and 238-239 sites of BBa_K1951008 and BBa_K1951009 (Protocol#3).
- From a starter of intermediate biobrick BBa_K1951004+ RBS BBa_K1951001, made culture at DO=0.2 until DO=0.6 and took a sample of 1uDO. Added 50µL beta mercatoethanol to the sample and congeled at -20°C to test protein production on a SDS page/Comassie.
Thursday, September 8th
- Starters of BBa_K1951008 + BbsI 215-216 position and starters of BBa_K1951008 + BbsI 238-239 position.
- Spreading of transduction in a E. coli W3110 strain using the phage P1 from the 09.05.2016 (500£µL cells + 2µL P1 or 10µL P1 or 10µL P1 or 50µL P1 or 100µL P1) (Protocol#8).
- Digestion of intermediate biobrick BBa_K1951004+ RBS BBa_K1951001 by EcoRI and SpeI and RBS BBa_K1951001+ RBS BBa_K1951002 by SpeI and PstI (Protocol#5).
- Ligation of the 2 previous digestions in pSB1C3 to make the final BBa_K1951011 (E/P) (Protocol#5).
- Transformation of the previous ligation in competent Tg1 (Protocol#2).
Friday, September 9th
- Derivatization of the sample cadA mutant complemented with BBa_K1951004 and cadA mutant complemented with BBa_K1951004+ RBS BBa_K1951001 .
- Reisolement of the knockout fliC mutant W3110 from the transformation of the 09.08.2016.
- Plasmid purification of the BBa_K1951008 + BbsI 215-216 position and starters of BBa_K1951008 + BbsI 238-239 position (Protocol#4).
- Digestion test by BBs1 and EcoRI using NEB buffer and electrophoresis analyse (Protocol#5).
- Colony PCR of BBa_K1951011 in pSB1C3 by amplification with different SLIC oligos FW and RV to see the differente constitutive biobrick. Biobrick is OK (Protocol#3).
Satursday, September 10th
- Swimming test of the knockout fliC mutant W3110 for 10 differents colonies and the W3110 WT as a positive temoin (Protocol#11).
- Starters of the fliC mutant W3110.
Sunday, September 11st
- Made competent of the fliC mutant W3110 strain from 5 differents colonies from 4 colonies which swam well (Protocol#1).
- Transformation of the previous competent with BBa_K1951008 and BBa_K1951009 (Protocol#2).
- Transformation of cadA mutant Keio strain with BBa_K1951011 (Protocol#2).
Monday, September 12rd
- Transformation of the fliC mutant W3110 competent with BBa_K1951008 (Protocol#2).
Tuesday, September 13th
- Swimming test of colonies obtained after transformation of the 09.12.2016. After 3h incubation at 37°C, swimming has been recovered (Protocol#11).
- Monitoring growth from differents strain to make comassie. From starters of fliC mutant W3110 BBa_K1951008, liC mutant W3110 BBa_K1951009, Tg1 BBa_K1951010, Tg1 WT (negative temoin), Tg1+pSB1C3 containing RFP (negative temoin), and cadA mutant BBa_K1951011, we made cultures at DO=0.2 in duplicate. At DO=0.6, half of the cultures has been induced with 0.02 arabinose for the biobrick containing an inductible promotor. Every hour, a sample of 1uDO was taken. After centrifugation at 5000g during 5min and removing of the supernatant, we added 50µL beta mercatoethanol and congeled at -20°C to test protein production on a SDS page/Comassie later.
- HPLC test of the fliC mutant complemented with BBa_K1951011 (Protocol#9).
Thursday, September 15
- Dillution of peptides oligos by 10000 (ex: 0.31mg/238µL/10000=ng/µL).
- Annealing 98°C during 2min and cool down during approximatly 3h
- Ligation of 5ng from the annealing mix for 100ng pSB1C3 digested by EcoRI and PstI (Protocol#5).
- Transformation in Tg1 (Protocol#2).