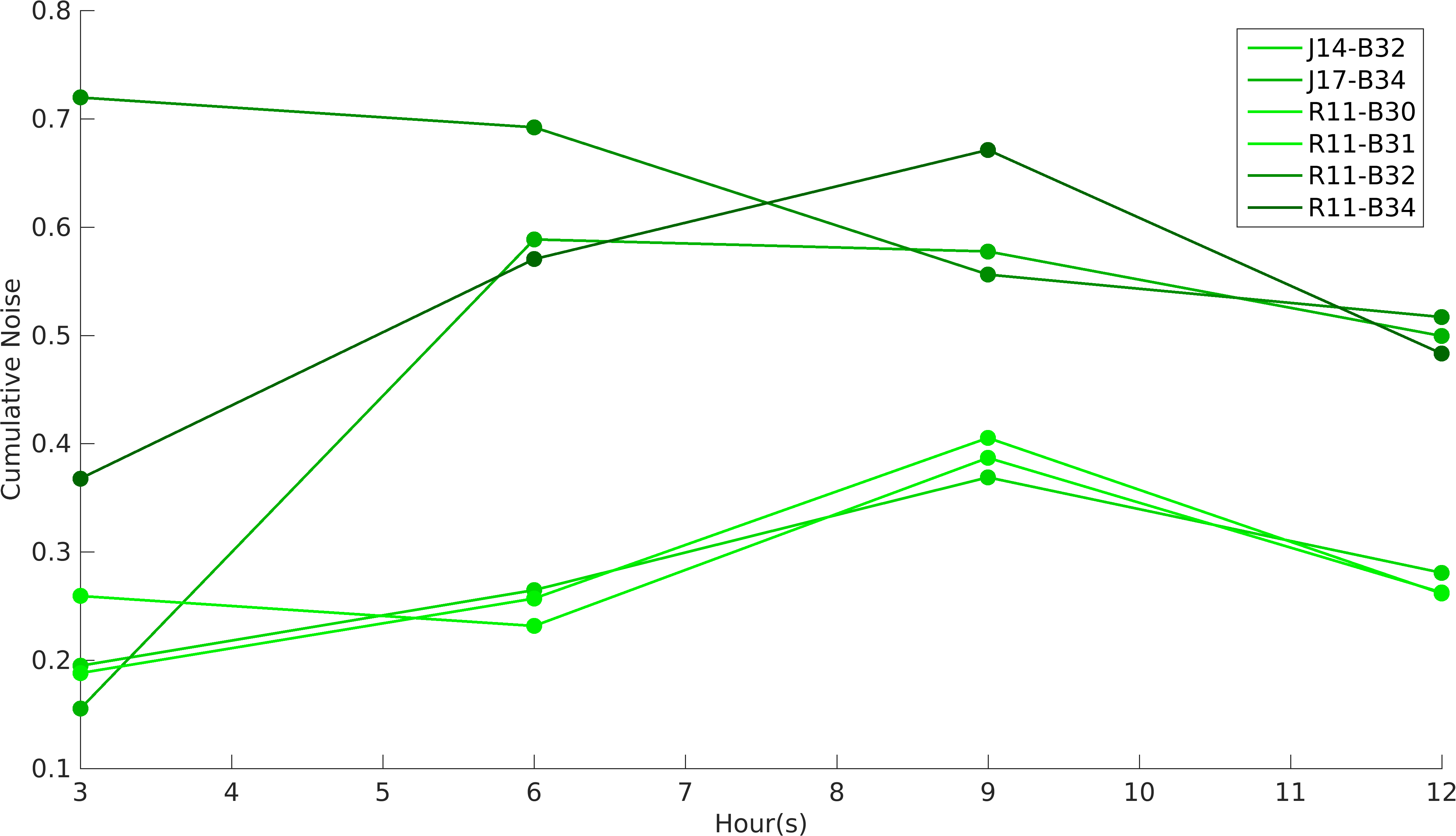
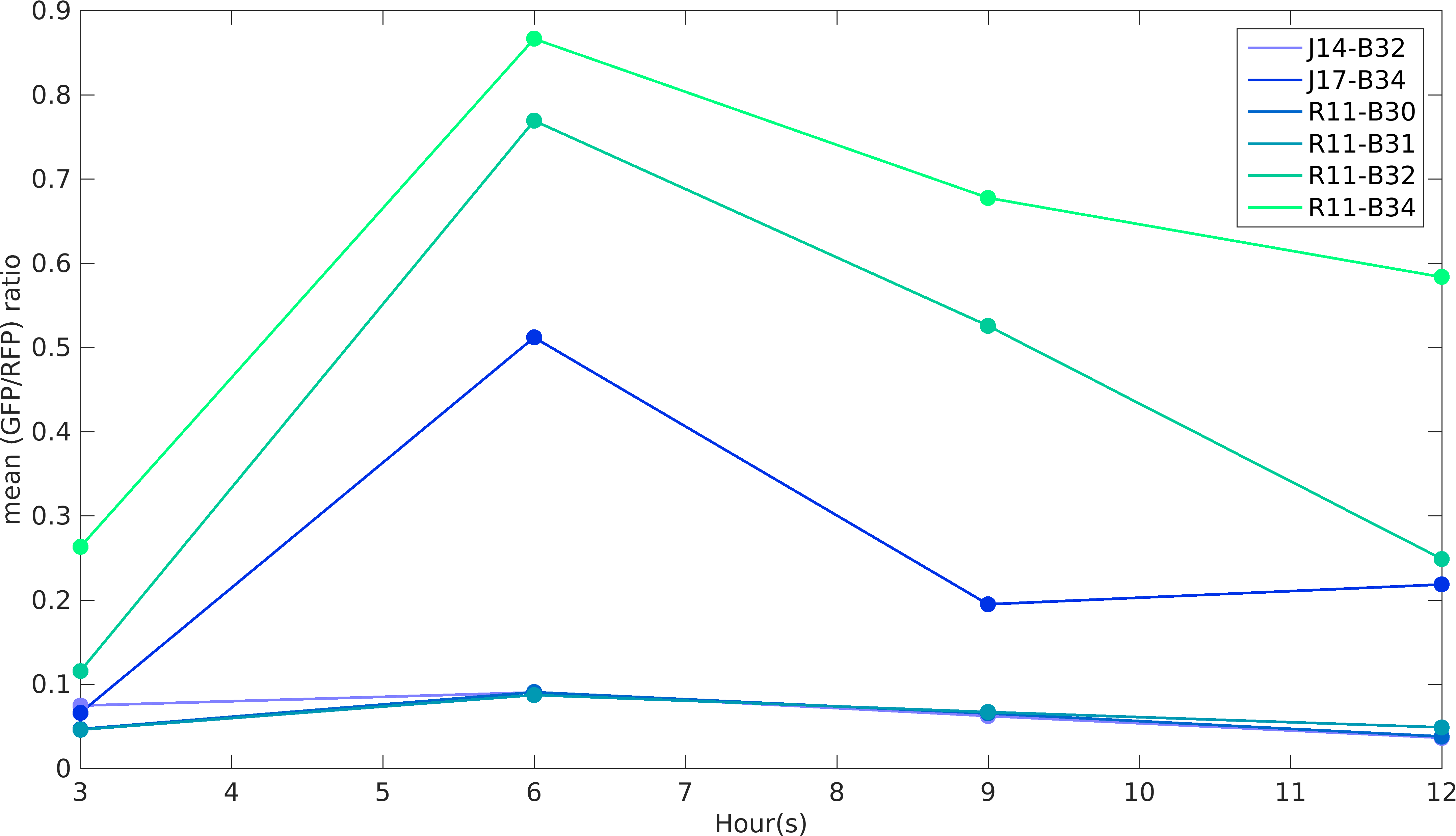
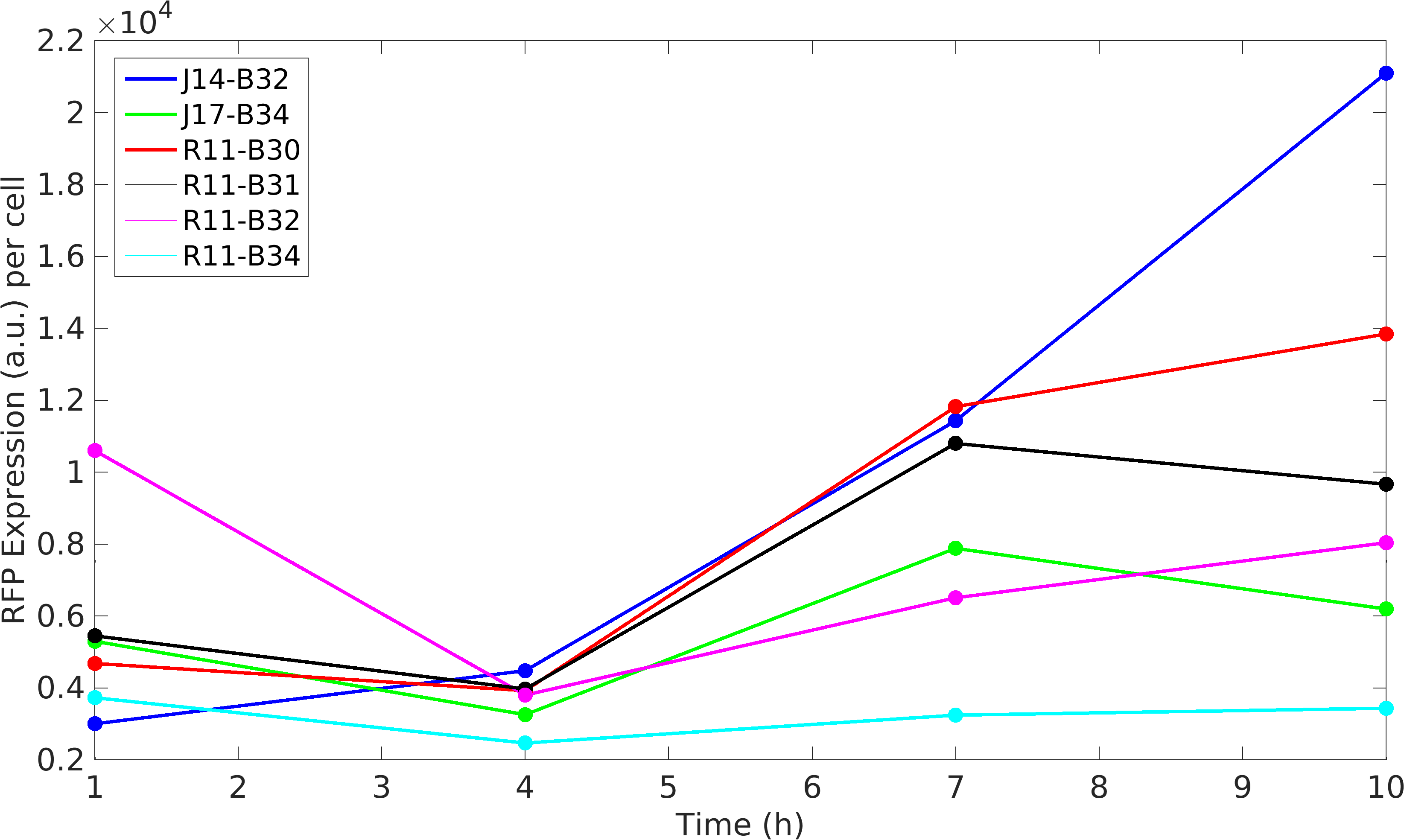
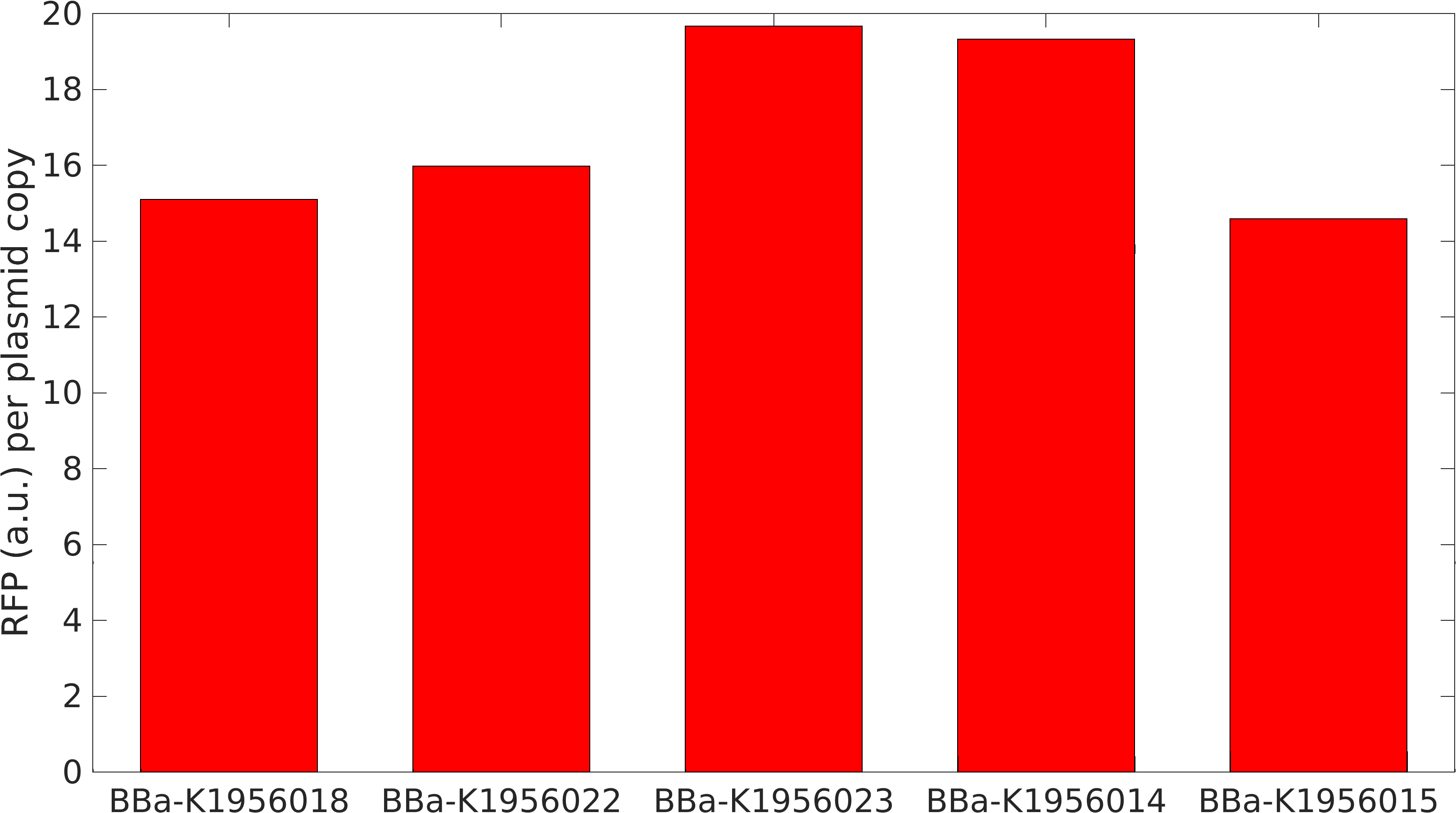
Noise in Devices
We analyzed GFP and RFP expression in all of our construct using flow cytometry. In our experiments, we have used [http://parts.igem.org/Part:BBa_R0011 R0011] and [http://parts.igem.org/Part:BBa_R0010 R0010] promoters to produce GFP and RFP proteins respectively. According to the characterization done by William & Mary iGEM 2015 team, R0010 is more noisy than R0011 promoter, while RBSs role in noise estimation was not taken into account in the characterization. While building upon William & Mary 2015's project, we were prompted to see the role of RBSs and different promoters on intrinsic noise. Also, we wanted to see how do the intrinsic noise of individual parts give rise to intrinsic noise of complex devices. Considering these objectives in mind, we designed our experiments. We worked with devices [http://parts.igem.org/Part:BBa_K1956022 K1956022], [http://parts.igem.org/Part:BBa_K1956023 K1956023], [http://parts.igem.org/Part:BBa_K1956014 K1956014], [http://parts.igem.org/Part:BBa_K1956015 K1956015], [http://parts.igem.org/Part:BBa_K1956017 K1956017] and [http://parts.igem.org/Part:BBa_K1956018 K1956018]. Following graph shows intrinsic noise values for every devices over different time points of it's growth curve. Our results conclude that increase in protein expression of devices leads to higher noise irrespective of the nature of RBS and promoter parts.
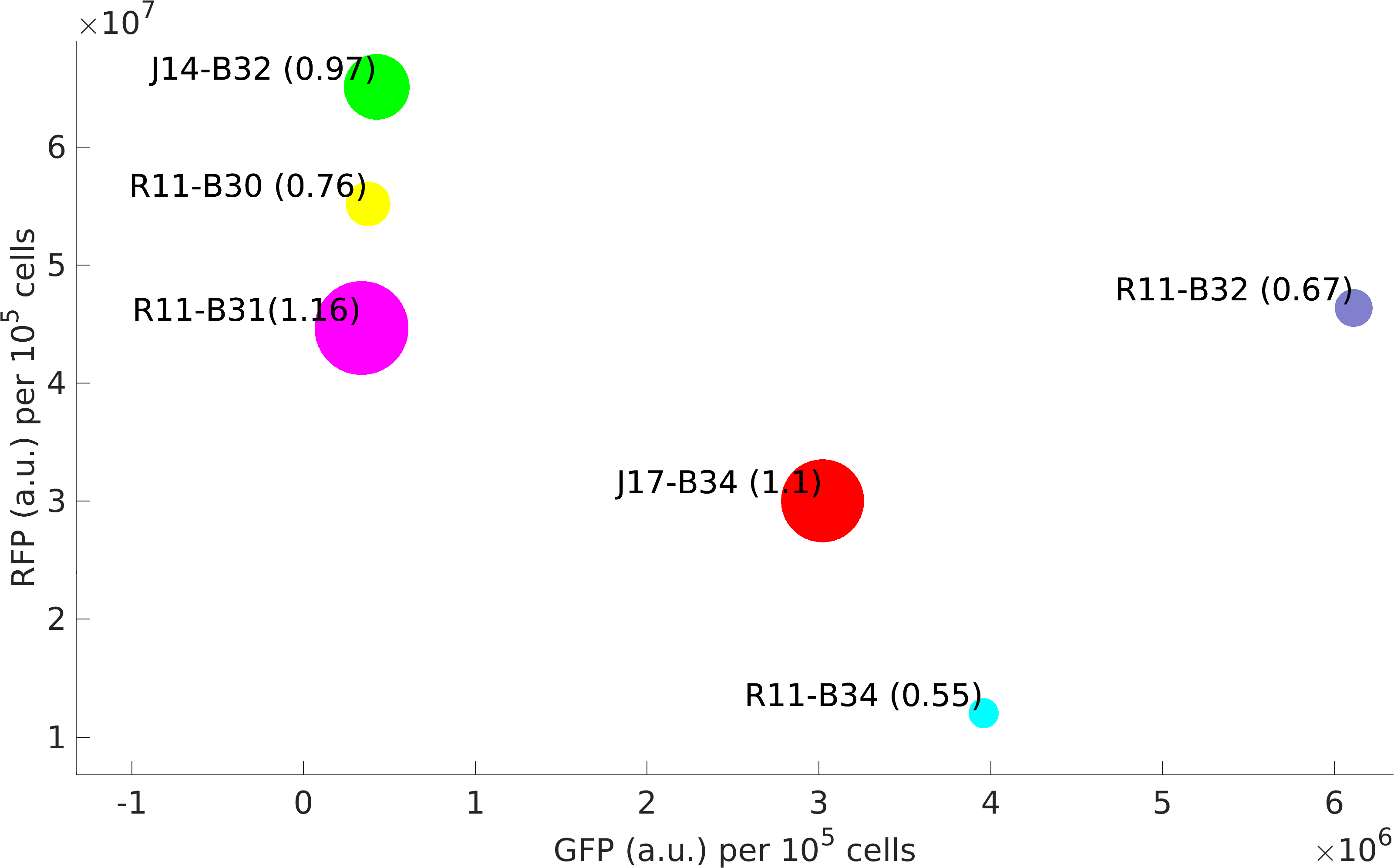
In this experiment of ours, we have done better characterization by using an internal control (RFP producing module) of the strength of promoter-RBS composite parts [http://parts.igem.org/Part:BBa_K1956001 K1956001], [http://parts.igem.org/Part:BBa_K1956002 K1956002], [http://parts.igem.org/Part:BBa_K1956003 K1956003], [http://parts.igem.org/Part:BBa_K1956001 K1956004], J23151-B0032 and J23117-B0034.
Further, we state the importance of internal control in our experiments. In our experiments, RFP producing module acts as internal control, we found huge variations in the RFP expression of our devices.
We isolated plasmid from E. coli DH5alpha cells with our devices. We found that RFP expression per plasmid copy is almost same in our devices. This not just signifies the importance of internal controls but also tell us that high protein expression reduces plasmid copy number in cells.
We also performed growth curve study and found that growth curve of cells with higher protein expressions (GFP, RFP) tend to have slower specific growth rate.
Also in our modelling section, we have estimated the strength and quantified the non-modularity of RBS part: B0030, B0032, B0034.
RIBOS
RIBOSON
Trigger RNA: A part of RNA molecule, which codes for [http://parts.igem.org/Part:BBa_E1010 RFP] protein.
Switch RNA: It was designed to repress the translation process by masking the RBS placed upstream of GFP protein coding part. After interacting with the RNA molecules of RFP gene, RIBOSON would allow the RBS to open up to bind to ribosomal machinery for protein synthesis to take place.
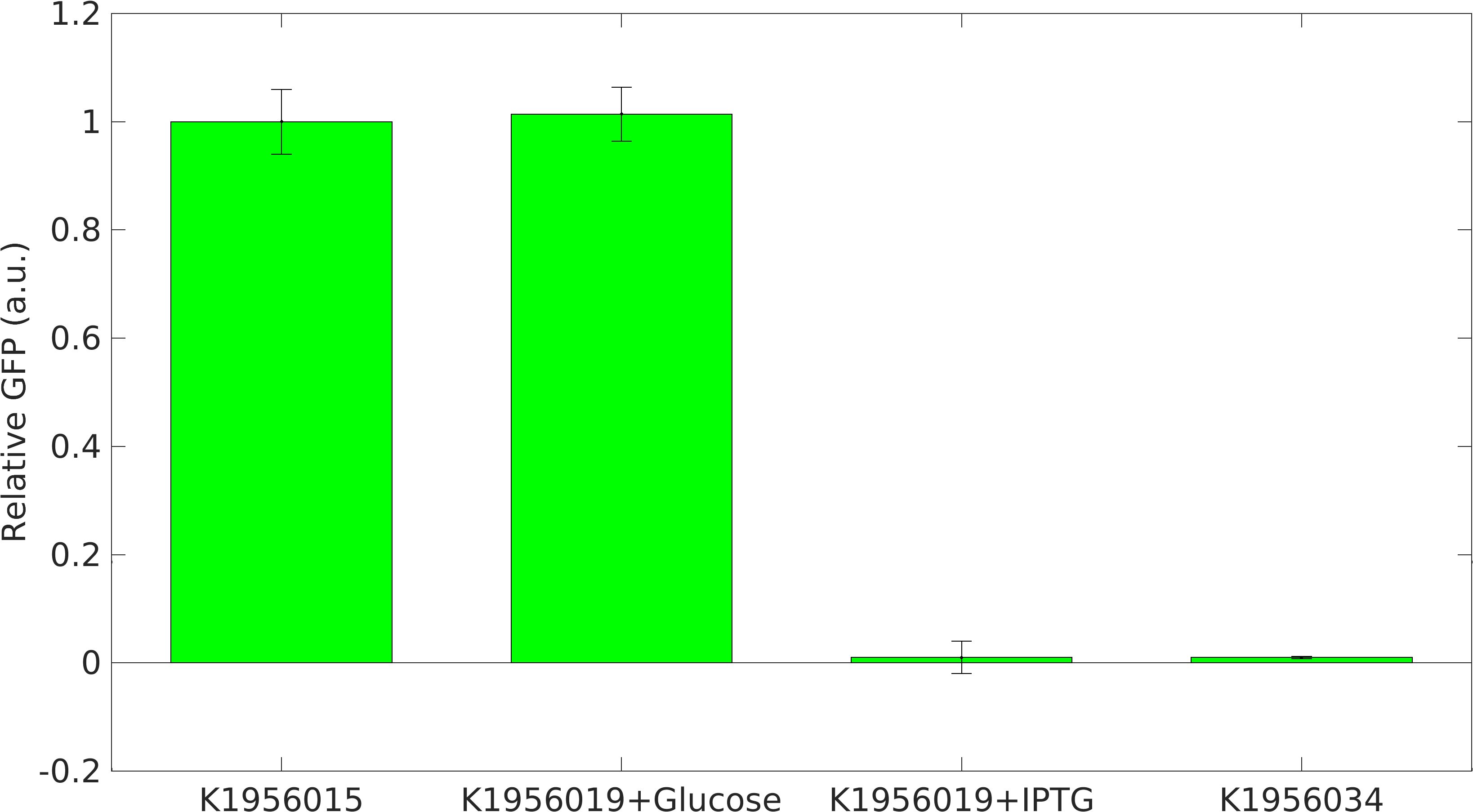
We performed our RIBOSON experiment in E. coli BL21(DE3). We transformed our cloned device [http://parts.igem.org/Part:BBa_K1956019 BBa_K1956019], which has RIBOSONrfp switch under [http://parts.igem.org/Part:BBa_J23100 J23100] constitutive promoter, upstream of [http://parts.igem.org/Part:BBa_E0040 GFP] coding part and RFP generator under [http://parts.igem.org/Part:BBa_R0011 lacI+pL] promoter.
We grew the cells in LB broth media + 40mM Glucose. After, 16 hrs of growth, we made two types of secondary culture: 1. Cells + LB broth + 40mM Glucose and 2. Cells + LB broth + 1mM IPTG.
Positive Control: Device [http://parts.igem.org/Part:BBa_K1956015 K1956015], which produces GFP and RFP proteins.
Negative Control: Device [http://parts.igem.org/Part:BBa_K1956034 K1956034], which produces lacI mRNA molecules.
RIBOSOFF
Trigger RNA: A part of RNA molecule, which codes for lacI protein.
Switch RNA: It was designed to activate the translation process by allowing the ribosomal machinery to bind to RBS, placed upstream of GFP protein coding part. After interacting with the RNA molecules of lacI gene, RIBOSOFF changes it's confirmation to mask the RBS and represses the translation process.
We performed our RIBOSOFF experiment in E. coli DH5alpha. We transformed our cloned device [http://parts.igem.org/Part:BBa_K1956020 BBa_K1956020], which has RIBOSOFFlacI switch under [http://parts.igem.org/Part:BBa_J23100 J23106] constitutive promoter, upstream of [http://parts.igem.org/Part:BBa_E0040 GFP] coding part and RFP generator under [http://parts.igem.org/Part:BBa_R0011 lacI+pL] promoter.
We prepared two types of cells: 1. [http://parts.igem.org/Part:BBa_K1956020 BBa_K1956020] in pSB1A2 + [http://parts.igem.org/Part:BBa_K1956034 BBa_K1956034] in pSB1K3, 2. [http://parts.igem.org/Part:BBa_K1956020 BBa_K1956020] only.
We grew the cells in LB broth media. After, 16 hrs of growth, we inoculated secondary cultures and collect samples at 12 hour of growth.
Positive Control: Device [http://parts.igem.org/Part:BBa_K1956015 K1956015], which produces GFP and RFP proteins.
Negative Control: Device [http://parts.igem.org/Part:BBa_K1956034 K1956034], which produces lacI mRNA molecules.