| Line 470: | Line 470: | ||
<!--End of outermost box--> | <!--End of outermost box--> | ||
<script type="text/javascript" src="https://2016.igem.org/Template:Peshawar/Javascript?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2016.igem.org/Template:Peshawar/Javascript?action=raw&ctype=text/javascript"></script> | ||
| + | <!-- Start of StatCounter Code for Default Guide --> | ||
| + | <script type="text/javascript"> | ||
| + | var sc_project=11100754; | ||
| + | var sc_invisible=1; | ||
| + | var sc_security="135c4f8d"; | ||
| + | var scJsHost = (("https:" == document.location.protocol) ? | ||
| + | "https://secure." : "http://www."); | ||
| + | document.write("<sc"+"ript type='text/javascript' src='" + | ||
| + | scJsHost+ | ||
| + | "statcounter.com/counter/counter.js'></"+"script>"); | ||
| + | </script> | ||
| + | <noscript><div class="statcounter"><a title="shopify | ||
| + | analytics ecommerce" href="http://statcounter.com/shopify/" | ||
| + | target="_blank"><img class="statcounter" | ||
| + | src="//c.statcounter.com/11100754/0/135c4f8d/1/" | ||
| + | alt="shopify analytics ecommerce"></a></div></noscript> | ||
| + | <!-- End of StatCounter Code for Default Guide --> | ||
</body> | </body> | ||
</html> | </html> | ||
Revision as of 06:55, 12 October 2016
Modelling (Under Construction)
$$\mathbf{[CO]+[CooA] \longleftrightarrow [(CooA)2CO]}$$
$$\mathbf{[(CooA)2CO] \longleftrightarrow [(CooA)2CO.Pr]}$$
$k_{on1} = \textrm{formation rate constant}$
$\gamma_1 = \textrm{degradation rate constant}$
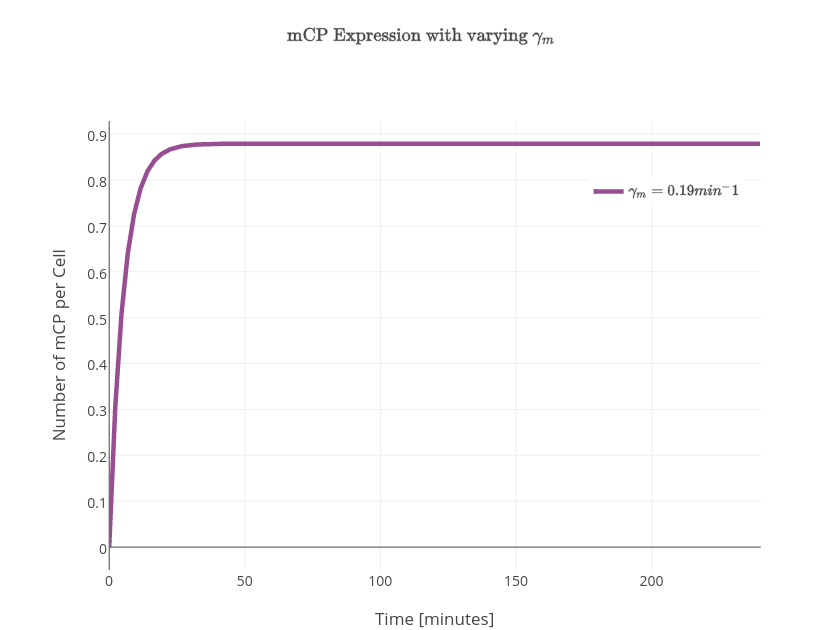
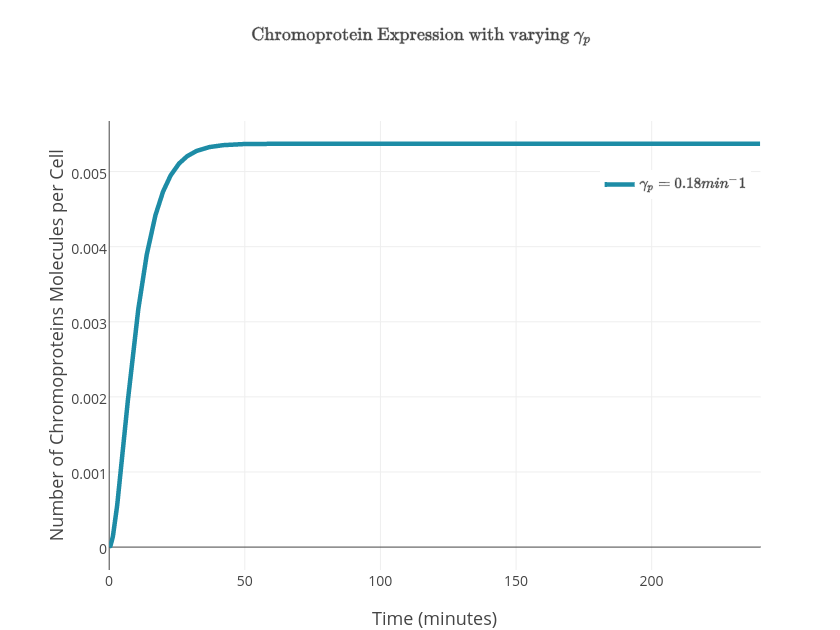
$P_{exp} = \textrm{probability of promoter to be activated}$ $$P_{exp} = \frac{\textrm{number of situations in which the promoter will be activated}}{\textrm{total number of situations}} = \frac{w}{z}$$ $$ w=\textrm{[(CooA)2CO.Pr]}=\frac{\textrm{[(CooA)2CO]}}{K_p}$$ $$\frac{d(mRNA)}{dt}=k_{on1}.P_{exp}-\gamma_1.[mRNA]$$ $$\textrm{Change in the concentration of the chromprotein} = \textrm{formation rate} - \textrm{degration rate}$$ $$ z=w+1=\frac{\textrm{[(CooA)2CO]}}{K_p}+1$$ $$\textrm{Total number of situations} = \textrm{Number of situations in which the promoter will be activated} - \textrm{Number of situations in which the promoter will not be acitvated}$$ $$P_{exp} = \frac{w}{z} = \frac{\frac{\textrm{[(CooA)2CO]}}{K_p}}{\frac{\textrm{[(CooA)2CO]}}{K_p}+1}$$ $$P_{exp} = \frac{\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} {1+\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} $$ $$[mRNA]=\frac{k_{on1}.P_{exp}}{\gamma_1}(1-e^{-\gamma_1t})$$ $$\frac{d\textrm{[CP]}}{dt} = k_{on2}.\textrm{[mRNA]}-\gamma_2\textrm{[CP]}$$ $$\textrm{[CP]} = k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1.\gamma_2}(1-e^{-\gamma_2t}) + k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1(\gamma_1-\gamma_2)}(e^{-\gamma_2t}-e^{-\gamma_1t})$$ $$\frac{d\textrm{[mCP]}}{dt} = k_{transcription}\textrm{[PYeaR]} - \gamma_m \textrm{[mCP]} $$ $$\frac{d\textrm{[CP]}}{dt}=k_{translation}\textrm{[mCP]}-\gamma_p\textrm{[CP]}$$ $$\frac{[\textrm{NO}^*]}{[\textrm{NO}_T]} = \frac{1}{1+\left(\frac{NsrR}{K_{NO}} \right)^n}$$ $$\textrm{[PYeaR]}activity = \frac{\beta}{1+ \left( \frac{\textrm{NO}^*}{K_{d(\textrm{NsrR})}} \right)}$$ $$\frac{d[\textrm{mCP}]}{dt} = \frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} } - \gamma_m [\textrm{mCP}]$$ $$K=\frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} }$$ $$\frac{d[\textrm{mCP}]}{dt} = K - \gamma_m [\textrm{mCP}]$$ $$[\textrm{mCP}]=\frac{K}{\gamma_m} - \frac{K}{\gamma_m}e^{-\gamma_mt}$$ $$[\textrm{CP}]=\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} e^{-\gamma_mt} - \left(\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} \right)e^{-\gamma_pt} $$
$\gamma_1 = \textrm{degradation rate constant}$
$P_{exp} = \textrm{probability of promoter to be activated}$ $$P_{exp} = \frac{\textrm{number of situations in which the promoter will be activated}}{\textrm{total number of situations}} = \frac{w}{z}$$ $$ w=\textrm{[(CooA)2CO.Pr]}=\frac{\textrm{[(CooA)2CO]}}{K_p}$$ $$\frac{d(mRNA)}{dt}=k_{on1}.P_{exp}-\gamma_1.[mRNA]$$ $$\textrm{Change in the concentration of the chromprotein} = \textrm{formation rate} - \textrm{degration rate}$$ $$ z=w+1=\frac{\textrm{[(CooA)2CO]}}{K_p}+1$$ $$\textrm{Total number of situations} = \textrm{Number of situations in which the promoter will be activated} - \textrm{Number of situations in which the promoter will not be acitvated}$$ $$P_{exp} = \frac{w}{z} = \frac{\frac{\textrm{[(CooA)2CO]}}{K_p}}{\frac{\textrm{[(CooA)2CO]}}{K_p}+1}$$ $$P_{exp} = \frac{\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} {1+\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} $$ $$[mRNA]=\frac{k_{on1}.P_{exp}}{\gamma_1}(1-e^{-\gamma_1t})$$ $$\frac{d\textrm{[CP]}}{dt} = k_{on2}.\textrm{[mRNA]}-\gamma_2\textrm{[CP]}$$ $$\textrm{[CP]} = k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1.\gamma_2}(1-e^{-\gamma_2t}) + k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1(\gamma_1-\gamma_2)}(e^{-\gamma_2t}-e^{-\gamma_1t})$$ $$\frac{d\textrm{[mCP]}}{dt} = k_{transcription}\textrm{[PYeaR]} - \gamma_m \textrm{[mCP]} $$ $$\frac{d\textrm{[CP]}}{dt}=k_{translation}\textrm{[mCP]}-\gamma_p\textrm{[CP]}$$ $$\frac{[\textrm{NO}^*]}{[\textrm{NO}_T]} = \frac{1}{1+\left(\frac{NsrR}{K_{NO}} \right)^n}$$ $$\textrm{[PYeaR]}activity = \frac{\beta}{1+ \left( \frac{\textrm{NO}^*}{K_{d(\textrm{NsrR})}} \right)}$$ $$\frac{d[\textrm{mCP}]}{dt} = \frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} } - \gamma_m [\textrm{mCP}]$$ $$K=\frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} }$$ $$\frac{d[\textrm{mCP}]}{dt} = K - \gamma_m [\textrm{mCP}]$$ $$[\textrm{mCP}]=\frac{K}{\gamma_m} - \frac{K}{\gamma_m}e^{-\gamma_mt}$$ $$[\textrm{CP}]=\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} e^{-\gamma_mt} - \left(\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} \right)e^{-\gamma_pt} $$
| Parameter | Description | Value |
|---|---|---|
| $[\textrm{PYeaR}]$ | Concentration of PYeaR | 1 copy/cell |
| $k_{transcription}$ | Rate of chromoprotein mRNA synthesis | 0.167$\textrm{min}^{-1}$ |
| $k_{translation}$ | Rate of chromoprotein synthesis | $0.0011\textrm{min}^{-1}$ |
| $\gamma_m$ | mRNA degradation rate | $0.19\textrm{min}^{-1}$ |
| $\gamma_p$ | Chromoprotein degradation rate | $0.18\textrm{min}^{-1}$ |
| $K_{\textrm{NO}}$ | Dissociation constant of NO | $0.12\textrm{min}^{-1}$ |
| $K_{\textrm{d(NsrR)}}$ | Dissociation constant of NsrR | 0.035 mM |
| n | Cooperativity of NO binding to NsrR | 2 |
| Header Cell 1 | Header Cell 2 | Header Cell 3 |
|---|---|---|
| Row 1 Cell 1 | Row 1 Cell 2 | |
| Row 2 Cell 1 | Row 2 Cell 2 | |
| Row 3 Cell 1 | Row 3 Cell 2 | |
| Row 4 Cell 1 | Row 4 Cell 2 |


 Institute of Integrative Biosciences,
Institute of Integrative Biosciences,

 +92 91 5860291
+92 91 5860291



