(→Proof of fonctionnality) |
|||
| Line 17: | Line 17: | ||
We investigated if DesA (Lysine decarboxylase) was fonctionnal, by measurement of cadaverine using HPLC with C18 column and proofed our biobrick is a producer of this protein and makes it fonctionnal. | We investigated if DesA (Lysine decarboxylase) was fonctionnal, by measurement of cadaverine using HPLC with C18 column and proofed our biobrick is a producer of this protein and makes it fonctionnal. | ||
| − | [[File:T--Aix-Marseille--result1.jpeg| | + | [[File:T--Aix-Marseille--result1.jpeg|770px|center|thumb|<u>On the right</u>, we tested the protein production using a SDS page and comassie in a <i>Escherichia coli</i> Tg1 strain after transformation with Bba_K1951011. On the right column, it has been observed the protein production without induction with 0.02% arabinose. On the left column, protein production was tested after induction. Results showed the production of the 4 proteins DesA, DesB, DesC and DesD, all involved in the desferrioxamine biosynthesis pathway. <u> On the left, </u> we had a look of the cadaverine production by the lysine decarboxylase DesA. Production has been detected by HPLC using C18 column after induction of the strain. <i>Escherichia coli</i> Tg1 strain was used as a wild type. Cadaverine production has been detected in this strain. In a Tg1 cadA mutant, cadaverine was also produced in a least quantity showing that an other pathway is responsible for the production of cadaverine. In the cadA mutant complemented by Bba_K1951004, the amount of cadaverine was recovered and even beyond the wild type production. Moreover, in the cadA mutant complemented by Bba_K1951011, the cadaverine level produced was even over the wild type and complemented Bba_K1951004 production ]] |
===In the futur=== | ===In the futur=== | ||
Result
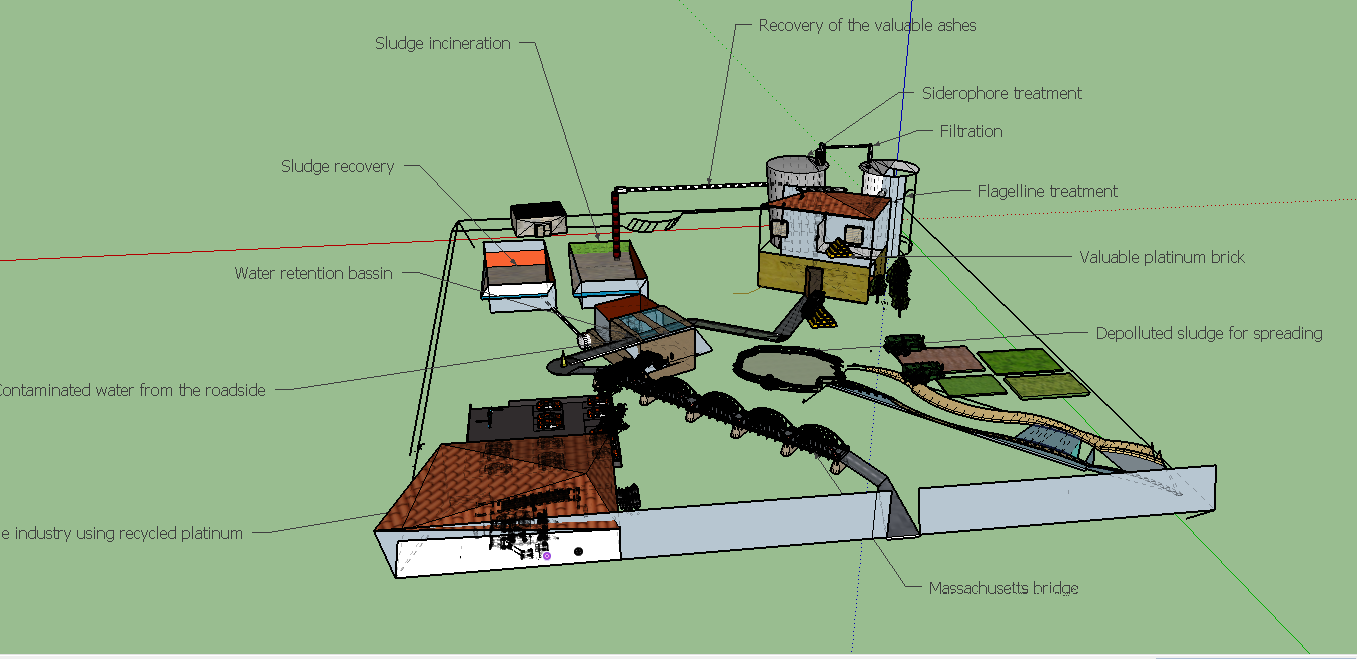
Our lab experience enabled us to create the final Biobrocks of our project which are a siderophore Desferrioxamine B producer and a flagellin producer.
Mobilisation result
Proof of protein production
Production of every enzymes involved in the pathway has been shown by a SDS page and Comassie Blue. We compared the production of proteins in different background :
- E. coli Tg1 strain without any plasmide
- E. coli Tg1 strain with pSB1C3 containing the RFP coding sequence
- E. coli Tg1 strain complemented with our biobrick Bba_K1951011 before and after induction. (you can observe the production of the 4 proteins on the figure below on the left; right : before induction, left : after induction)
Proof of fonctionnality
We investigated if DesA (Lysine decarboxylase) was fonctionnal, by measurement of cadaverine using HPLC with C18 column and proofed our biobrick is a producer of this protein and makes it fonctionnal.

In the futur
We plan to test the whole pathway viability in the first hand. In a second hand, the potential adsorption of platium by our siderophore would be investigate in a rich metal medium.
Biosorption result
Proof of swimming recovery
We have made a biobrick BbaK1951008 ables to produce Flagellin (FliC protein of the flagellum). In the aim to test the flagellin integrity, we made a fliC mutant in a E.coli W3110 strain by transduction using phage P1 (protocol available on our website). fliC mutant has been complemented by our biobrick Bba_K1951008. On the following figure, we investigated if the complemented fliC mutant recovered swimming after complementation. To do this, we did a swimming test using soft gelose. WT W3110 strain was well swimming after 3 hours incubation at 37°C. However, FliC mutant wasn't able to swim whereas fliC mutant complemented recover the swimming ability, making a proof that our biobrick is production a fonctionnal flagellin protein.

Microscopie of the flagellum
We analysed the fliC mutant complemented by BbaK1951008 using electronic microscopy. This tools allowed us to observe the flagellum integrity recovered and to obtain phenotypic image of our work.
In the futur
We plan to make a proof of concept of the platinum absorption to the surface of the flagellin. In a second time, we want to finish the insertion of the restriction site to allow the cassette insertion ( peptide absorbing specifically precious metal) and test their affinity.
Improvement of FliC E. coli BBa_K342000
This biobrick has been improoved from a previous one designed by Glasgow 2014 team. Please find the link of this biobrick below : http://parts.igem.org/Part:BBa_K1463601
Instead of promotors Bba_J23106 and Bba_J23116, we used strong promoter, strong RBS combination for high expression levels of the flagellin. By the combination of Bba_K880005 and Bba_K1951005, we made a high flagellin expression vector able to recover swimming.
Project achievement in a table
Futur plan for our project
Now that every biobricks have been created and are available, make proofs of concept could allow to envisage investigation about possibilities of an industrial application.
The biologic production of high and specific adsorber could reduce environnemental impact of the metal production and its cost while renew essential precious metals like platinum, for a sustainable production.
Project Achievements