(Prototype team page) |
|||
| Line 1: | Line 1: | ||
| − | {{Hong_Kong_UCCKE}} | + | {{Hong_Kong_UCCKE/CSS/reset.css}} {{Hong_Kong_UCCKE/CSS/bootstrap.css}} {{Hong_Kong_UCCKE/CSS/style.css}} {{Hong_Kong_UCCKE/CSS/latofonts.css}} {{Hong_Kong_UCCKE/CSS/font-awesome.css}} {{Hong_Kong_UCCKE/CSS/jcarousel.responsive.css}} {{Hong_Kong_UCCKE/CSS/bootstrap.toc.css}} |
<html> | <html> | ||
| + | <link rel="stylesheet" href="https://2016.igem.org/Team:Leiden/libraries/font-awesome-4-6-3/css/font-awesome-min?action=raw&ctype=text/css" type="text/css" /> | ||
| + | <meta name="viewport" content="width=device-width, initial-scale=1"> | ||
| + | <div class="wholehead"> | ||
| − | < | + | </html> |
| − | + | {{Hong_Kong_UCCKE/crazyheader}} | |
| − | < | + | <html> |
| − | + | <div class="header" id="header"> | |
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col-sm-12 page_header-text"> | ||
| + | <div> | ||
| + | <h1 data-toc-skip>UCCKE / Composite Part</h1> | ||
| + | <p>In this page, you can get an overall image of our composite parts</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-sm-12" style="text-align:center;"> | ||
| + | <a href="#normalsec" class="no-text-dec"> | ||
| + | <i class="fa fa-angle-down header-down" aria-hidden="true" style="font-size:40px; margin-bottom:10px;"></i> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| − | + | </div> | |
</div> | </div> | ||
| + | </div> | ||
| + | <style> | ||
| + | .normalsec h2 { | ||
| + | padding-top:0; | ||
| + | } | ||
| + | </style> | ||
| + | <div class="normalsec" id="normalsec" style="text-align:left !important;"> | ||
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col-sm-9 col-xs-12"> | ||
| + | <h2 style="margin-top:0; padding-top:0;">Introduction</h2> | ||
| + | <p style="text-align:left !important; font-size: 15px !important;"> | ||
| + | |||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <div class="col-sm-4 col-xs-12"> | ||
| + | <script src="https://2016.igem.org/Template:Hong_Kong_UCCKE/JS/bootstraptoc?action=raw&ctype=text/javascript" type="text/javascript"></script> | ||
| + | </div> | ||
| + | <div class="col-sm-9 col-xs-12" style="text-align:left !important;"> | ||
| + | <h2 style="margin-top:0; padding-top:0;">K1889007</h2> | ||
| + | <p style="text-align:left !important; font-size: 15px !important;"><b>Express amino acid deaminase under constitutive promoter</b><br><br></p> | ||
| + | <p style="text-align:left !important; font-size: 15px !important;">K1889007 is a gene coding for an amino acid deaminase gene. It is derived from Proteus mirabilis. It functions to turn phenylalanine into phenylpyruvic acid by removing its amino group, coming from the enzyme family that catalyzes the breakdown of amino acids into alpha-keto acids. In our experiment, this gene is used to turn phenylalanine, a naturally occurring amino acid in E. coli, into phenylpyruvic acid. The phenylpyruvic acid then acts as an attractant to C. elegans.<br><br></p> | ||
| − | < | + | <img src="https://static.igem.org/mediawiki/parts/2/20/Team-Hong_Kong_UCCKE_2016_img24rfjui45t98u5.jpeg" style="width:100%; max-width:600px;"><br><br> |
| + | <p style="text-align:left !important; font-size: 15px !important;">The sequence of the gene was referenced from an article from Journal of Bacteriology, titled “Proteus mirabilis amino acid deaminase: cloning, nucleotide sequence, and characterization of aad.” The DNA sequence was slightly modified to allow its chemical synthesis by IDT while still retaining the original amino acid sequence.<br><br></p> | ||
| + | <p style="text-align:left !important; font-size: 15px !important;">When put in a composite part, the amino acid deaminase gene can produce phenylpyruvic acid in the transformed cell. Phenylpyruvic acid was then used in the droplet test assay. The phenylpyruvic acid were put in opposing poles while distilled water served as a control. (fig. 2) By calculating the index of the C. elegans dispersed on the plate, we obtained the following result. (fig. 3)<br><br></p> | ||
| − | < | + | <img src="https://static.igem.org/mediawiki/parts/9/94/Team-hong_kong-uccke-img2.png" style="width:100%; max-width:600px;"><br><br> |
| − | + | ||
| − | < | + | |
| − | < | + | <img src="https://static.igem.org/mediawiki/parts/f/f4/Team-hong-kong-uccke-img3.png" style="width:100%; max-width:600px;"><br><br> |
| + | <p style="text-align:left !important; font-size: 15px !important;">As seen by the above data, the higher the concentration of concentration, the higher the index. This reflects that the number of worms concentrated around the phenylpyruvic acid droplet increases with its concentration. This proves that the phenylpyruvic acid indeed attracts C. elegans. As such, phenylpyruvic acid is an attractant. Hence, when the gene is expressed in the transformed cell, it can produce the same attractant effect.<br><br></p> | ||
| + | </div> | ||
| + | <div class="col-sm-4 col-xs-12"></div> | ||
| − | |||
| − | |||
| − | |||
| + | <style> | ||
| + | nav[data-toggle='toc'] .nav .active .nav { | ||
| + | display: none; | ||
| + | } | ||
| + | |||
| + | #toc { | ||
| + | background-color: initial; | ||
| + | border: none; | ||
| + | } | ||
| + | </style> | ||
| + | <nav id="toc"></nav> | ||
| + | <script> | ||
| + | $(function () { | ||
| + | var navSelector = '#toc'; | ||
| + | var $myNav = $(navSelector); | ||
| + | Toc.init($myNav); | ||
| + | $('body').scrollspy({ | ||
| + | target: navSelector | ||
| + | }); | ||
| + | }); | ||
| + | </script> | ||
| + | <script> | ||
| + | var fixmeTop = $('.normalsec').offset().top; | ||
| + | $(window).scroll(function () { | ||
| + | if (window.innerWidth < 768) { | ||
| + | $('#toc').css({ | ||
| + | position: 'relative' | ||
| + | , display: 'none' | ||
| + | }); | ||
| + | } else { | ||
| + | var scrollTop = $(".normalsec").offset().top - $(window).scrollTop(); | ||
| + | var currentScroll = $(window).scrollTop(); | ||
| + | if (currentScroll >= fixmeTop) { | ||
| + | $('#toc').css({ | ||
| + | position: 'relative' | ||
| + | , top: scrollTop * -1 | ||
| + | }); | ||
| + | } else { | ||
| + | $('#toc').css({ | ||
| + | position: 'static' | ||
| + | }); | ||
| + | } | ||
| + | } | ||
| + | }); | ||
| + | </script> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| + | </html> | ||
| + | <html> | ||
| + | <div class="container"> | ||
| + | </html> | ||
| + | {{Hong_Kong_UCCKE/tourbefore|Notebook|Notebook|Basic_Part|Basic_Part}} | ||
| + | <html> | ||
</div> | </div> | ||
| − | |||
</html> | </html> | ||
| + | {{Hong_Kong_UCCKE/heyfooter}} | ||
| + | {{Hong_Kong_UCCKE/illbeback}} | ||
Revision as of 12:01, 16 October 2016
Introduction
K1889007
Express amino acid deaminase under constitutive promoter
K1889007 is a gene coding for an amino acid deaminase gene. It is derived from Proteus mirabilis. It functions to turn phenylalanine into phenylpyruvic acid by removing its amino group, coming from the enzyme family that catalyzes the breakdown of amino acids into alpha-keto acids. In our experiment, this gene is used to turn phenylalanine, a naturally occurring amino acid in E. coli, into phenylpyruvic acid. The phenylpyruvic acid then acts as an attractant to C. elegans.

The sequence of the gene was referenced from an article from Journal of Bacteriology, titled “Proteus mirabilis amino acid deaminase: cloning, nucleotide sequence, and characterization of aad.” The DNA sequence was slightly modified to allow its chemical synthesis by IDT while still retaining the original amino acid sequence.
When put in a composite part, the amino acid deaminase gene can produce phenylpyruvic acid in the transformed cell. Phenylpyruvic acid was then used in the droplet test assay. The phenylpyruvic acid were put in opposing poles while distilled water served as a control. (fig. 2) By calculating the index of the C. elegans dispersed on the plate, we obtained the following result. (fig. 3)
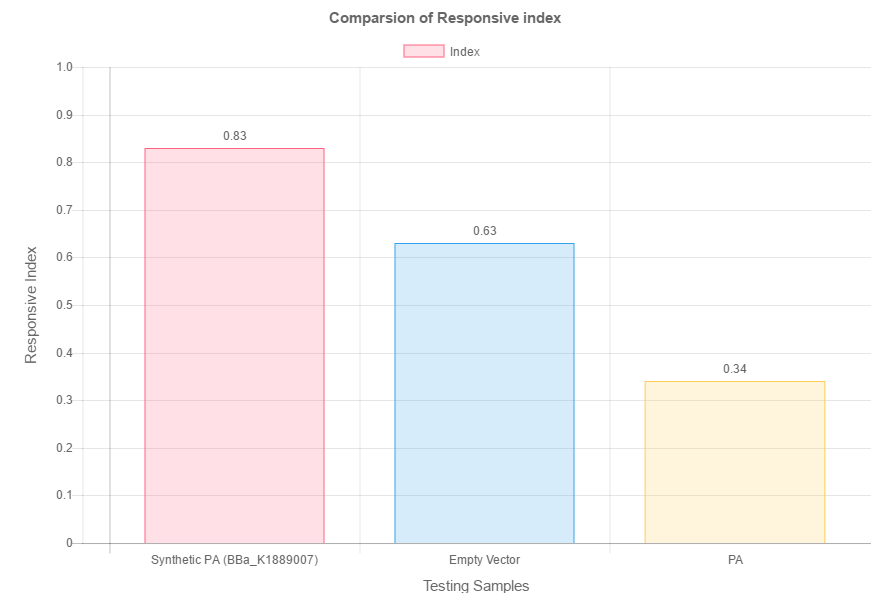

As seen by the above data, the higher the concentration of concentration, the higher the index. This reflects that the number of worms concentrated around the phenylpyruvic acid droplet increases with its concentration. This proves that the phenylpyruvic acid indeed attracts C. elegans. As such, phenylpyruvic acid is an attractant. Hence, when the gene is expressed in the transformed cell, it can produce the same attractant effect.


