Mariealice (Talk | contribs) (→Part Composition:) |
Mariealice (Talk | contribs) (→Lysine decarboxylase DesA :) |
||
| Line 12: | Line 12: | ||
===='''Lysine decarboxylase DesA''' :==== | ===='''Lysine decarboxylase DesA''' :==== | ||
| − | Lysine descarboxylase DesA from ''Streptomyces coelicolor'' is an enzyme from the lyase family that converts lysine | + | Lysine descarboxylase DesA from ''Streptomyces coelicolor'' is an enzyme from the lyase family that converts lysine into cadaverin. The enzyme releases the carbonyl group of the lysin amino acid. Cadaverine (1,5-diaminopentane) is a primary diamine which alkaline environment. Acidic pH and an anaerobic environment both induce the synthesis of Lysine decarboxylase. |
In bacteriology, this enzyme is sought through the middle of Moeller lysine or medium lysine Taylor. | In bacteriology, this enzyme is sought through the middle of Moeller lysine or medium lysine Taylor. | ||
| − | We registered the original sequence of this subpart in the iGEM registry of standard parts ([http://parts.igem.org/Part:BBa_K1951000 BBa_K1951000]). We optimized our sequence for ''E. coli'' and ordered the synthesis by addition of an | + | We registered the original sequence of this subpart in the iGEM registry of standard parts ([http://parts.igem.org/Part:BBa_K1951000 BBa_K1951000]). We optimized our sequence for ''E. coli'' and ordered the synthesis by addition of an inducible promoter. |
===[http://parts.igem.org/Part:BBa_K1951011 BBa_K1951011] : Desferrioxamine B producer pathway <i> Streptomyces coelicolor </i> producer=== | ===[http://parts.igem.org/Part:BBa_K1951011 BBa_K1951011] : Desferrioxamine B producer pathway <i> Streptomyces coelicolor </i> producer=== | ||
Contents
- 1 Mobilisation by a Siderophore parts
- 2 Biosorption parts
Composite part
Mobilisation by a Siderophore parts
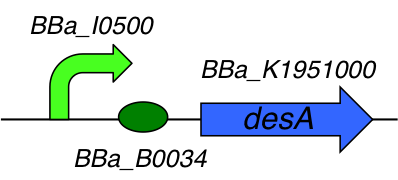
[http://parts.igem.org/Part:BBa_K1951004 BBa_K1951004] : DesA producer
Part Composition:
This part is composed of 3 subparts :
- [http://parts.igem.org/Part:BBa_I0500 I0500] : pARA/araC inducible promoter
- [http://parts.igem.org/Part:BBa_B0034 B0034] : Ribosome Binding Site
- [http://parts.igem.org/Part:BBa_K1951000 K1951000] : Lysine decarboxylase
Lysine decarboxylase DesA :
Lysine descarboxylase DesA from Streptomyces coelicolor is an enzyme from the lyase family that converts lysine into cadaverin. The enzyme releases the carbonyl group of the lysin amino acid. Cadaverine (1,5-diaminopentane) is a primary diamine which alkaline environment. Acidic pH and an anaerobic environment both induce the synthesis of Lysine decarboxylase. In bacteriology, this enzyme is sought through the middle of Moeller lysine or medium lysine Taylor.
We registered the original sequence of this subpart in the iGEM registry of standard parts ([http://parts.igem.org/Part:BBa_K1951000 BBa_K1951000]). We optimized our sequence for E. coli and ordered the synthesis by addition of an inducible promoter.
[http://parts.igem.org/Part:BBa_K1951011 BBa_K1951011] : Desferrioxamine B producer pathway Streptomyces coelicolor producer
Biosorption parts
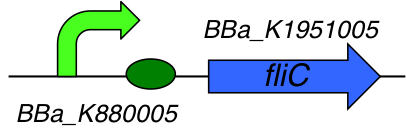
[http://parts.igem.org/Part:BBa_K1951008 BBa_K1951008] : FliC E. coli producer
Part Composition:
This part is a composite part composed of 2 Biobricks :
- [http://parts.igem.org/Part:BBa_K1951005 BBa_K1951005] : Flagellin C
- [http://parts.igem.org/Part:BBa_K880005 BBa_K880005] : Strong promoter, strong RBS combination
Flagellin C from Escherichia coli
Flagellin C (FliC) protein from Escherichia coli strain is the main protein constitutive of the flagelar filament and is involved to promote bacterial swimming. This sequence is conserved in many bacterial strains. It has been demonstrated that flagellin has the ability to adsorb precious metal such as platinum or gold. We made a FliC mutant by transduction using phage P1 in a E. Coli W3110 strain. Then we have complemented the fliC mutant W3110 with [http://parts.igem.org/Part:BBa_K1951008 BBa_K1951008] and performed a swimming test for every background. The result has shown that swimming was recovered into the complemented fliC mutant W3110
Part Assembly:
The subparts were assembled using standard BioBrick Assembly.
Biobrick amelioration
This biobrick has been improved from a previous one designed by Glasgow 2014 team. Please find the link of this biobrick below :
http://parts.igem.org/Part:BBa_K1463601
Instead of [http://parts.igem.org/Part:BBa_J23106 BBa_J23106] and [http://parts.igem.org/Part:BBa_J23116 BBa_J23116], we used strong promoter, strong RBS combination for high expression levels of the flagellin. By the combination of [http://parts.igem.org/Part:BBa_Bba_K880005 BBa_Bba_K880005] and [http://parts.igem.org/Part:BBa_K1951005 BBa_K1951005], we made a high flagellin expression vector able to recover swimming.
[http://parts.igem.org/Part:BBa_K1941009 BBa_K1941009] : FliC Desulfovibrio vulgaris producer
General
Flagellin C (FliC) protein from Desulfovibrio vulgaris strain is the main protein constitutive of the flagellum filament and is involved to promote bacterial swimming. This sequence is conserved in many bacterial strains as the capacity of swimming given by the flagellum confers a great selective advantage.

Metal Biosorption Capacity
It has been demonstrated that Flagellin has the ability to adsorb precious metal on its surface such as platinum, palladium gold [2][3] and this was important for our project Highway to platinum
Immune response capacity
The propensity of the immune response to flagellin may be explained by two facts:
- Flagellin is an extremely abundant protein in flagellated bacteria.
- There exists a specific innate immune receptor that recognizes flagellin, Toll-like receptor 5 (TLR5). [4]
[http://parts.igem.org/Part:BBa_K19410010 BBa_K1941010] : CsgA Escherichia coli producer
CsgA is the major and structural subunit of the curli fimbriae. Curli are coiled surface structures that assemble preferentially at growth temperatures below 37 degrees Celsius. Curli are the major proteinaceous component of a complex extracellular matrix produced by many Enterobacteriaceae. Curli were first discovered in the late 1980s on Escherichia coli strains that caused bovine mastitis, and have since been implicated in many physiological and pathogenic processes of E. coli and Salmonella spp. Curli fibers are involved in adhesion to surfaces, cell aggregation, and biofilm formation. Curli also mediate host cell adhesion and invasion, and they are potent inducers of the host inflammatory response. The biobrick contains a strong promotor.