| (7 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<div class="pageSection"> | <div class="pageSection"> | ||
<h2>Notebook</h2> | <h2>Notebook</h2> | ||
| − | + | <p><span style="color:blue; text-decoration;">'''Group 1: Cloning'''</span></p> | |
| + | <p><span style="color:green; text-decoration;">'''Group 2: Protein and Conjugation/Transformation into <i> Shewanella oneidensis'''</i></span></p> | ||
| + | <p><span style="color:orange; text-decoration;">'''Group 3: Electrochemistry'''</span></p> | ||
<div class="timeline_vline "> | <div class="timeline_vline "> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 14;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 1 4.07-8.07 | Week 1 4.07-8.07 | ||
| Line 12: | Line 14: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | ||
| − | <p> | + | [[File:T--NRP-UEA-Norwich--IMG_1872.JPG|130px]] |
| − | At the start of the week, team members completed a safety induction for the lab. The first experiment used the iGEM standard RFP BioBrick (BBa_J04450) to transform E. coli. For our planned outreach activity, seven coloured | + | <p> <span style="color:blue; text-decoration;"> |
| + | At the start of the week, team members completed a safety induction for the lab. The first experiment used the iGEM standard RFP BioBrick (BBa_J04450) to transform <i>E. coli</i>. For our planned outreach activity, seven coloured BioBricks from the iGEM 2016 distribution kit were identified. <i>E. coli</i> was transformed with these BioBricks, and then plated to determine which cells had been transformed successfully. </span> | ||
</p> | </p> | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
| Line 22: | Line 25: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 13;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 2 11.07-15.07 | Week 2 11.07-15.07 | ||
| Line 28: | Line 31: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | <p> | + | <p> <span style="color:blue; text-decoration;"> Successful transformants were selected and used to inoculate LB media, these samples were to be used for the children's Summer School. Consequently, they were mini prepped to obtain the plasmid DNA. Alongside this, the PCR protocol was optimised by experimenting with annealing temperatures. The <i>HydC/Fdh, HydA, HydB</i> C- and N-terminus genes and RFP BioBricks (Bba_J04450) were digested with <i>EcoR1</i> and <i>Pst1</i>. Samples were run on a gel to observe complete digestion and then extracted and purified. </span> |
| − | + | ||
</p> | </p> | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
| Line 37: | Line 39: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 12;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 3 18.07-22.07 | Week 3 18.07-22.07 | ||
| Line 43: | Line 45: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | <p> | + | <p><span style="color:blue; text-decoration;"> For the Summer School: Blue, Pink, RFP (Red Fluorescent Protein) and GFP (Green Fluorescent Protein) colonies were inoculated. Various drawing utensils were tested on plates to determine which one would be best for the children to use. After speaking to the student leader of UCC (<span class="plainlinks">[https://2016.igem.org/Team:UCC_Ireland iGEM Cork]</span>) it was decided to go with cotton buds. For our main project, the cleaned-up PCR products were run on a gel to check the PCR amplification was successful. 2.5µL DMSO was added to the <i>HyaC</i> N- and C-terminus DNA to minimise non-specific amplification, but did not make any difference. The FeFe gene inserts were digested along with RFP to obtain the BioBrick vector. Gel extraction and purification of the gene inserts yielded low concentrations, these were repeated. </span> </p> |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 52: | Line 52: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 11;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 4 25.07-29.07 | Week 4 25.07-29.07 | ||
| Line 58: | Line 58: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | ||
| − | <p> | + | [[File:T--NRP-UEA-Norwich--IMG_1958.JPG|130px]] |
| − | + | <p> <span style="color:blue; text-decoration;"> Vials of the ‘paint’ and cotton buds were prepared for the human practices activity. GFP, RFP, Pink and Blue were inoculated and grown overnight to use the following day for the activity. The PCR amplification was repeated for FeFe gene inserts. These were then digested, extracted and purified from the agarose gel. FeFe genes inserts were ligated with the BioBrick vector. The ligations reactions were set up with different ratios of vector and insert (1:1 and 1:3) and incubated overnight. These were transformed into <i>α-select</i> Gold Efficiency <i>E.coli</i> cells by heat shock the following day and plated out. Four plates showed colonies and successful ligations i.e. <i>HydC/Fdh</i> 1:1, <i>HydC/Fdh</i> 1:3, <i>HydA</i> 1:3 and <i>HydB</i> C-terminus 1:3. Digestion reactions were set up for the NiFe genes: <i>HyaC</i>, <i>HyaA</i> and <i>HyaB</i> N- and C-terminus followed by ligations. </span> </p> | |
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 68: | Line 67: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 10;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 5 1.08-5.08 | Week 5 1.08-5.08 | ||
| Line 74: | Line 73: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--pcr.jpg|130px]] | |
| − | <p> | + | <p> <span style="color:blue; text-decoration;">The successful colonies for <i>HydC/Fdh, HydA</i> and <i>HydB</i> C-terminus were inoculated in 10ml LB overnight. Following mini-prep plasmid purification, PCR confirmed correct ligations. The gel results showed unexpected sizes for some of the samples and suggested that some of the samples had been mis-labelled. DNA from new overnight cultures of <i>HydC/Fdh, HydA</i> and <i>HydB</i> C-terminus were obtained by mini-prep and a restriction digest was carried out to verify the results. Digests of <i>HydB</i> N-terminus, <i>HyaC, HyaA, HyaB</i> N-terminus and <i>HyaB</i> C-terminus were set up, however following gel purification concentrations were too low, thus transformations did not yield results. </span></p> |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 84: | Line 81: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 9;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 6 8.08-12.08 | Week 6 8.08-12.08 | ||
| Line 90: | Line 87: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | <p> | + | <p><span style="color:blue; text-decoration;"> We repeated the PCR amplification of FeFe units along with NiFe genes inserts. With new primers for <i>HyaB</i> N- and C-terminus gene insert. Gel showed successful amplification of FeFe gene inserts and <i>HyaB</i> N-terminus. The FeFe subunits were digested alongside samples of RFP DNA. Samples were extracted and purified from the gel, however nanodrop results showed extremely low concentration for <i>HydA</i>. It was decided this would be repeated. Afterwards, <i>HydA</i> and <i>HydB</i> N-terminus were ligated into the pSB1C3 vector and frozen for later transformation. More J04450 was inoculated and mini-prepped as a source of vector. <i>HydC/Fdh, HydB</i> C-terminus and <i>HyaA</i> was successfully ligated into pSB1C3 and digest confirmed a mix-up in sample labels however bands did correspond to correct gene inserts. </span> </p> |
| − | + | <p><span style="color:green; text-decoration;"> A practice tri-parental conjugation trial was conducted following protocol, using samples obtained from a PhD student in the lab </span></p> | |
| − | + | <p><span style="color:orange; text-decoration;"> We conducted experiments to practise growing bacteria for creating anaerobic growth curves and to practise using the gas chromatograph. We grew wildtype MR-1 and double knock out (∆<i>HydABC,HyaABC</i>) <i>Shewanella</i> strains in anaerobic conditions for 24 hours in hungate tubes then analysed the headspace gases using gas chromatography to calculate the hydrogen yield. </span> </p> | |
| − | <p> | + | |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 102: | Line 96: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 8;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 7 15.08-19.08 | Week 7 15.08-19.08 | ||
| Line 108: | Line 102: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | <p> | + | <p><span style="color:blue; text-decoration;">Due to the uncertainty of the identity of these potential BioBricks, <i>HydC/Fdh, HydB</i> N-terminus and <i>HyaA</i> plasmid DNA samples were sent for DNA sequencing confirmation. The results proved that these samples were successful BioBricks, but that samples had been mislabelled, samples were subsequently correctly labelled. The frozen samples of ligated <i>HydA</i> and <i>HydB</i> N-terminus were transformed successfully into <i>α-select E. coli</i> and incubated overnight. A colony from each was used to inoculate LB, incubated overnight and mini-prepped the following day. </span></p> |
| − | + | <p><span style="color:green; text-decoration;">We conducted a practice arabinose induced protein expression trial using samples from a PhD student in the lab. SDS-PAGE gels were set up and used to practice analysing protein expression. The tri-parental conjugation of RFP (J04450) Biobrick into <i>S. oneidensis</i> was set up for the following week. </span></p> | |
| − | + | <p><span style="color:orange; text-decoration;"> We grew MR1 and ∆<i>HydABC,HyaABC</i> <i>Shewanella</i> strains in anaerobic conditions for 24 hours in hungate tubes and analysed the amount of hydrogen produced using gas chromatography. This was to serve as a control value for our later experiments. </span> </p> | |
| − | <p> | + | |
| − | + | ||
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 121: | Line 111: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 7;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 8 22.08-26.08 | Week 8 22.08-26.08 | ||
| Line 127: | Line 117: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--IMG_2229.JPG|130px]] | |
| − | <p> | + | <p><span style="color:blue; text-decoration;">Golden Gate Cloning of FeFe Hydrogenase subunits was now possible. The use of the arabinose inducible pBAD vector allowed controlled expression of the FeFe Hydrogenase. Additionally, using pBAD allowed us to get around the reported issues of <i>S.oneidensis</i> not being compatible with pSB1C3. <span class="plainlinks"> [https://2008.igem.org/Team:Harvard iGEM Harvard 2008] </span> and <span class="plainlinks">[https://2015.igem.org/Team:NTU-Singapore NTU Singapore 2015] </span> have reported similar issues in conjugating BioBricks into <i>S.oneidensis</i> due to conflicts in origin of replication. This was also reported in a paper “Replication of plasmids with the p15A origin in <i>Shewanella putrefaciens</i>” (Myers and Myers ,1997). The Golden Gate cloning for FeFe hydrogenase (<i>Fdh/HydC, HydA</i> and <i>HydB</i> N-terminus) cluster was successful, allowing us to characterise our construct in <i>S.oneidensis</i>. Only <i>HyaC</i> and <i>HyaB</i> N-terminus were PCR amplified successfully, and cloned into pSB1C3. </span></p> |
| − | + | <p><span style="color:green; text-decoration;">Tri-parental conjugation of RFP (J04450) and GFP (K584001) BioBricks into S. oneidensis was attempted but was unsuccessful. </span> </p> | |
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | |||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 141: | Line 127: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 6;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 9 29.08-2.09 | Week 9 29.08-2.09 | ||
| Line 147: | Line 133: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | <p> | + | <p><span style="color:blue; text-decoration;">We obtained the <i>HydABC</i> FeFe hydrogenase gene cluster in our expression vector: pBAD, from the Golden Gate cloning. This was then transformed into <i>E.coli</i> and the overnight culture mini-prepped to obtain DNA, which was sent off for sequencing. Some of the remaining DNA was run on a gel against the pBAD vector to observe correct ligation. <i>HydABC</i> in our expression vector, pBAD, was transformed back into <i>α-select E.coli</i> for more cells. The NiFe gene inserts were transformed and plated, but was unsuccessful. Additional RFP digests showed high amounts of smearing, thus an overnight digest was done and used to ligate the NiFe gene inserts.</span></p> |
| − | + | <p><span style="color:green; text-decoration;">Wild Type <i>S. oneidensis</i> MR-1, ∆<i>HydABC,HyaABC</i> (NiFe and FeFe hydrogenase double knockout), <i>LS498</i> (FeFe hydrogenase knockout) and <i>pRK2013</i> (<i>E.coli</i> helper strain) were inoculated in preparation for transforming the plasmid containing <i>HydABC</i> into <i>S. oneidensis</i>. Triparental conjugation was then performed to transform 8A5 (pBAD vector with <i>HydABC</i> C-terminally tagged) and 9A5 (pBAD vector with <i>HydABC</i> N-terminally tagged) into all three strains of <i>S. oneidensis</i>. </span></p> | |
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 159: | Line 141: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 5;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 10 5.09-9.09 | Week 10 5.09-9.09 | ||
| Line 165: | Line 147: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--IMG_2122.JPG|130px]] | |
| − | <p> | + | <p><span style="color:blue; text-decoration;">Ligated <i>HyaA</i> and <i>HyaB</i> N-terminus did not yield any colonies. Stocks of DNA were made by inoculating and mini-prepping more samples of the <i>HydABC</i>. </span> </p> |
| − | + | <p><span style="color:green; text-decoration;">The triparental conjugation was continued from last week, however there were no <i>S. oneidensis</i> colonies containing the plasmid. Attempted electroporation to deliver the plasmid with <i>HydABC</i> into <i>S. oneidensis</i>. The first attempt did not yield any results, but the second attempt gave colonies of <i>S.oneidensis</i> MR-1 and ∆<i>HydABC,HyaABC</i>strains <i>S. oneidensis</i> with both 8A5 and 9A5. </span></p> | |
| − | + | <p><span style="color:orange; text-decoration;">We grew∆<i>HydABC,HyaABC</i> and wildtype MR-1 strains for 24 hours in anaerobic conditions. We then used an electrochemical cell setup to analyse the activation of the hydrogenase enzymes, and the difference between the two strains. This would then serve as a control for later experiments. </span></p> | |
| − | <p> | + | |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 178: | Line 157: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 4;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 11 12.09-16.09 | Week 11 12.09-16.09 | ||
| Line 184: | Line 163: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--IMG_2291.JPG|130px]] | |
| − | <p> | + | <p><span style="color:blue; text-decoration;">Attempted to start again with NiFe catalytic subunit cloning. PCR amplified with new primers and digested with a new source of <i>EcoR1</i> and <i>Pst1</i> as digests with our enzymes showed high amounts of smearing. Digested gel showed a band for <i>HyaB</i> N-terminus but not for <i>HyaB</i> C-terminus. Ligations, at ratios of 1:3, 1:5 and 1:7, were set up to be transformed the following week. Alongside <i>HyaB</i> N- and C-terminus, it was decided to excise <i>HydABC</i> out of the pBAD vector and ligate that into pSB1C3.</span></p> |
| − | + | <p><span style="color:green; text-decoration;">Mini-prepped colonies of the electroporation were sent off for sequencing. Results showed that only wild type <i>S. oneidensis</i> MR-1 has successfully taken up the <i>HydABC</i> C-terminally tagged i.e. 8A5, construct. Triparental conjugation and electroporation was continued with other strains but did not yield results. Protein expression trials (induced with 0mM, 1mM and 10mM arabinose) of the WT with 8A5, <i>E.coli</i> with 8A5 and 9A5 were performed using SDS-PAGE and western blot techniques. </span></p> | |
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 197: | Line 172: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="left" style="z-index: | + | <div class="timeline_event_header" data-side="left" style="z-index: 3;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 12 19.09-23.09 | Week 12 19.09-23.09 | ||
| Line 203: | Line 178: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--IMG_2316.JPG|130px]] | |
| − | <p> | + | <p><span style="color:blue; text-decoration;">NiFe Hydrogenase subunits and <i>HydABC</i> were ligated and transformed into <i>E.coli</i> cells. All plates had colonies, however the digested gel samples showed that our insert was not in them this was most likely due to incomplete digestion of Bba_J04450. The DNA concentrations of <i>HyaB</i> C- and N-terminus were low. Thus, we decided to order new DNA, in order to re-attempt cloning of <i>HyaB</i> C- and N-terminus into pSB1C3 </span></p> |
| − | + | <p><span style="color:green; text-decoration;">The triparental conjugations and electroporations from the previous week did not give any positive results. Protein expression trials (induced with 0mM, 1mM and 10mM arabinose) using SDS-PAGE and Western Blot were repeated for Mr1:<i>HydABC</i> and <i>E.coli</i> with 8A5 and 9A5. </span></p> | |
| − | + | ||
| − | <p> | + | |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 216: | Line 187: | ||
</div> | </div> | ||
<div class="timeline_bubble"> | <div class="timeline_bubble"> | ||
| − | <div class="timeline_event_header" data-side="right" style="z-index: | + | <div class="timeline_event_header" data-side="right" style="z-index: 2;"> |
<div class="timeline_event_header_container"> | <div class="timeline_event_header_container"> | ||
Week 13 26.09-30.09 | Week 13 26.09-30.09 | ||
| Line 222: | Line 193: | ||
<div class="timeline_event_container"> | <div class="timeline_event_container"> | ||
<div class="timeline_event_inner"> | <div class="timeline_event_inner"> | ||
| − | + | [[File:T--NRP-UEA-Norwich--IMG_2291.JPG|130px]] | |
| − | <p> | + | <p><span style="color:blue; text-decoration;">The digests of RFP were repeated to obtain a source of vector for new ligations for NiFe gene inserts and <i>HydABC</i> cluster. There were transformed into <i>E.coli</i> cells. All plates showed the ligations were successful as colonies had grown. The colonies were used to inoculate LB and mini prepped to perform a diagnostic colony PCR. Results showed correct ligations of all NiFe gene inserts, yielding more BioBricks. </span></p> |
| − | + | ||
| − | + | ||
<div class="close_timeline_event"></div> | <div class="close_timeline_event"></div> | ||
</div> | </div> | ||
| Line 231: | Line 200: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="timeline_bubble"> | |
| + | <div class="timeline_event_header" data-side="left" style="z-index: 1;"> | ||
| + | <div class="timeline_event_header_container"> | ||
| + | Week 14 3.10-8.10 | ||
| + | </div> | ||
| + | <div class="timeline_event_container"> | ||
| + | <div class="timeline_event_inner"> | ||
| + | <p> <span style="color:orange; text-decoration;">We grew wildtype MR-1 and MR-1 FeFe overexpression strains (our modified strain i.e. <i>HydABC</i> ligated into pBAD by Golden Gate cloning) in anaerobic conditions with varying concentrations of arabinose added. This showed the difference in expression between arabinose concentrations and also the effect of adding our FeFe <i>HydABC</i> hydrogenase cluster into the bacteria. This data could also be compared to MR1 without arabinose.</span></p> | ||
| + | <div class="close_timeline_event"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
{{NRP-UEA-Norwich-footer}} | {{NRP-UEA-Norwich-footer}} | ||
Latest revision as of 21:28, 18 October 2016
Notebook
Group 1: Cloning
Group 2: Protein and Conjugation/Transformation into Shewanella oneidensis
Group 3: Electrochemistry
Week 1 4.07-8.07
At the start of the week, team members completed a safety induction for the lab. The first experiment used the iGEM standard RFP BioBrick (BBa_J04450) to transform E. coli. For our planned outreach activity, seven coloured BioBricks from the iGEM 2016 distribution kit were identified. E. coli was transformed with these BioBricks, and then plated to determine which cells had been transformed successfully.
Week 2 11.07-15.07
Successful transformants were selected and used to inoculate LB media, these samples were to be used for the children's Summer School. Consequently, they were mini prepped to obtain the plasmid DNA. Alongside this, the PCR protocol was optimised by experimenting with annealing temperatures. The HydC/Fdh, HydA, HydB C- and N-terminus genes and RFP BioBricks (Bba_J04450) were digested with EcoR1 and Pst1. Samples were run on a gel to observe complete digestion and then extracted and purified.
Week 3 18.07-22.07
For the Summer School: Blue, Pink, RFP (Red Fluorescent Protein) and GFP (Green Fluorescent Protein) colonies were inoculated. Various drawing utensils were tested on plates to determine which one would be best for the children to use. After speaking to the student leader of UCC (iGEM Cork) it was decided to go with cotton buds. For our main project, the cleaned-up PCR products were run on a gel to check the PCR amplification was successful. 2.5µL DMSO was added to the HyaC N- and C-terminus DNA to minimise non-specific amplification, but did not make any difference. The FeFe gene inserts were digested along with RFP to obtain the BioBrick vector. Gel extraction and purification of the gene inserts yielded low concentrations, these were repeated.
Week 4 25.07-29.07
Vials of the ‘paint’ and cotton buds were prepared for the human practices activity. GFP, RFP, Pink and Blue were inoculated and grown overnight to use the following day for the activity. The PCR amplification was repeated for FeFe gene inserts. These were then digested, extracted and purified from the agarose gel. FeFe genes inserts were ligated with the BioBrick vector. The ligations reactions were set up with different ratios of vector and insert (1:1 and 1:3) and incubated overnight. These were transformed into α-select Gold Efficiency E.coli cells by heat shock the following day and plated out. Four plates showed colonies and successful ligations i.e. HydC/Fdh 1:1, HydC/Fdh 1:3, HydA 1:3 and HydB C-terminus 1:3. Digestion reactions were set up for the NiFe genes: HyaC, HyaA and HyaB N- and C-terminus followed by ligations.
Week 5 1.08-5.08
The successful colonies for HydC/Fdh, HydA and HydB C-terminus were inoculated in 10ml LB overnight. Following mini-prep plasmid purification, PCR confirmed correct ligations. The gel results showed unexpected sizes for some of the samples and suggested that some of the samples had been mis-labelled. DNA from new overnight cultures of HydC/Fdh, HydA and HydB C-terminus were obtained by mini-prep and a restriction digest was carried out to verify the results. Digests of HydB N-terminus, HyaC, HyaA, HyaB N-terminus and HyaB C-terminus were set up, however following gel purification concentrations were too low, thus transformations did not yield results.
Week 6 8.08-12.08
We repeated the PCR amplification of FeFe units along with NiFe genes inserts. With new primers for HyaB N- and C-terminus gene insert. Gel showed successful amplification of FeFe gene inserts and HyaB N-terminus. The FeFe subunits were digested alongside samples of RFP DNA. Samples were extracted and purified from the gel, however nanodrop results showed extremely low concentration for HydA. It was decided this would be repeated. Afterwards, HydA and HydB N-terminus were ligated into the pSB1C3 vector and frozen for later transformation. More J04450 was inoculated and mini-prepped as a source of vector. HydC/Fdh, HydB C-terminus and HyaA was successfully ligated into pSB1C3 and digest confirmed a mix-up in sample labels however bands did correspond to correct gene inserts.
A practice tri-parental conjugation trial was conducted following protocol, using samples obtained from a PhD student in the lab
We conducted experiments to practise growing bacteria for creating anaerobic growth curves and to practise using the gas chromatograph. We grew wildtype MR-1 and double knock out (∆HydABC,HyaABC) Shewanella strains in anaerobic conditions for 24 hours in hungate tubes then analysed the headspace gases using gas chromatography to calculate the hydrogen yield.
Week 7 15.08-19.08
Due to the uncertainty of the identity of these potential BioBricks, HydC/Fdh, HydB N-terminus and HyaA plasmid DNA samples were sent for DNA sequencing confirmation. The results proved that these samples were successful BioBricks, but that samples had been mislabelled, samples were subsequently correctly labelled. The frozen samples of ligated HydA and HydB N-terminus were transformed successfully into α-select E. coli and incubated overnight. A colony from each was used to inoculate LB, incubated overnight and mini-prepped the following day.
We conducted a practice arabinose induced protein expression trial using samples from a PhD student in the lab. SDS-PAGE gels were set up and used to practice analysing protein expression. The tri-parental conjugation of RFP (J04450) Biobrick into S. oneidensis was set up for the following week.
We grew MR1 and ∆HydABC,HyaABC Shewanella strains in anaerobic conditions for 24 hours in hungate tubes and analysed the amount of hydrogen produced using gas chromatography. This was to serve as a control value for our later experiments.
Week 8 22.08-26.08
Golden Gate Cloning of FeFe Hydrogenase subunits was now possible. The use of the arabinose inducible pBAD vector allowed controlled expression of the FeFe Hydrogenase. Additionally, using pBAD allowed us to get around the reported issues of S.oneidensis not being compatible with pSB1C3. iGEM Harvard 2008 and NTU Singapore 2015 have reported similar issues in conjugating BioBricks into S.oneidensis due to conflicts in origin of replication. This was also reported in a paper “Replication of plasmids with the p15A origin in Shewanella putrefaciens” (Myers and Myers ,1997). The Golden Gate cloning for FeFe hydrogenase (Fdh/HydC, HydA and HydB N-terminus) cluster was successful, allowing us to characterise our construct in S.oneidensis. Only HyaC and HyaB N-terminus were PCR amplified successfully, and cloned into pSB1C3.
Tri-parental conjugation of RFP (J04450) and GFP (K584001) BioBricks into S. oneidensis was attempted but was unsuccessful.
Week 9 29.08-2.09
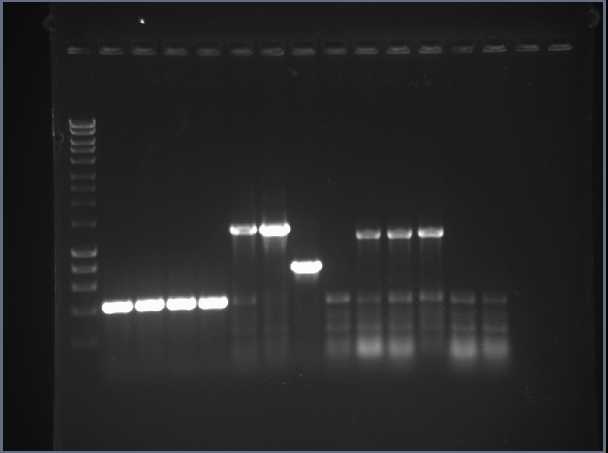
We obtained the HydABC FeFe hydrogenase gene cluster in our expression vector: pBAD, from the Golden Gate cloning. This was then transformed into E.coli and the overnight culture mini-prepped to obtain DNA, which was sent off for sequencing. Some of the remaining DNA was run on a gel against the pBAD vector to observe correct ligation. HydABC in our expression vector, pBAD, was transformed back into α-select E.coli for more cells. The NiFe gene inserts were transformed and plated, but was unsuccessful. Additional RFP digests showed high amounts of smearing, thus an overnight digest was done and used to ligate the NiFe gene inserts.
Wild Type S. oneidensis MR-1, ∆HydABC,HyaABC (NiFe and FeFe hydrogenase double knockout), LS498 (FeFe hydrogenase knockout) and pRK2013 (E.coli helper strain) were inoculated in preparation for transforming the plasmid containing HydABC into S. oneidensis. Triparental conjugation was then performed to transform 8A5 (pBAD vector with HydABC C-terminally tagged) and 9A5 (pBAD vector with HydABC N-terminally tagged) into all three strains of S. oneidensis.
Week 10 5.09-9.09
Ligated HyaA and HyaB N-terminus did not yield any colonies. Stocks of DNA were made by inoculating and mini-prepping more samples of the HydABC.
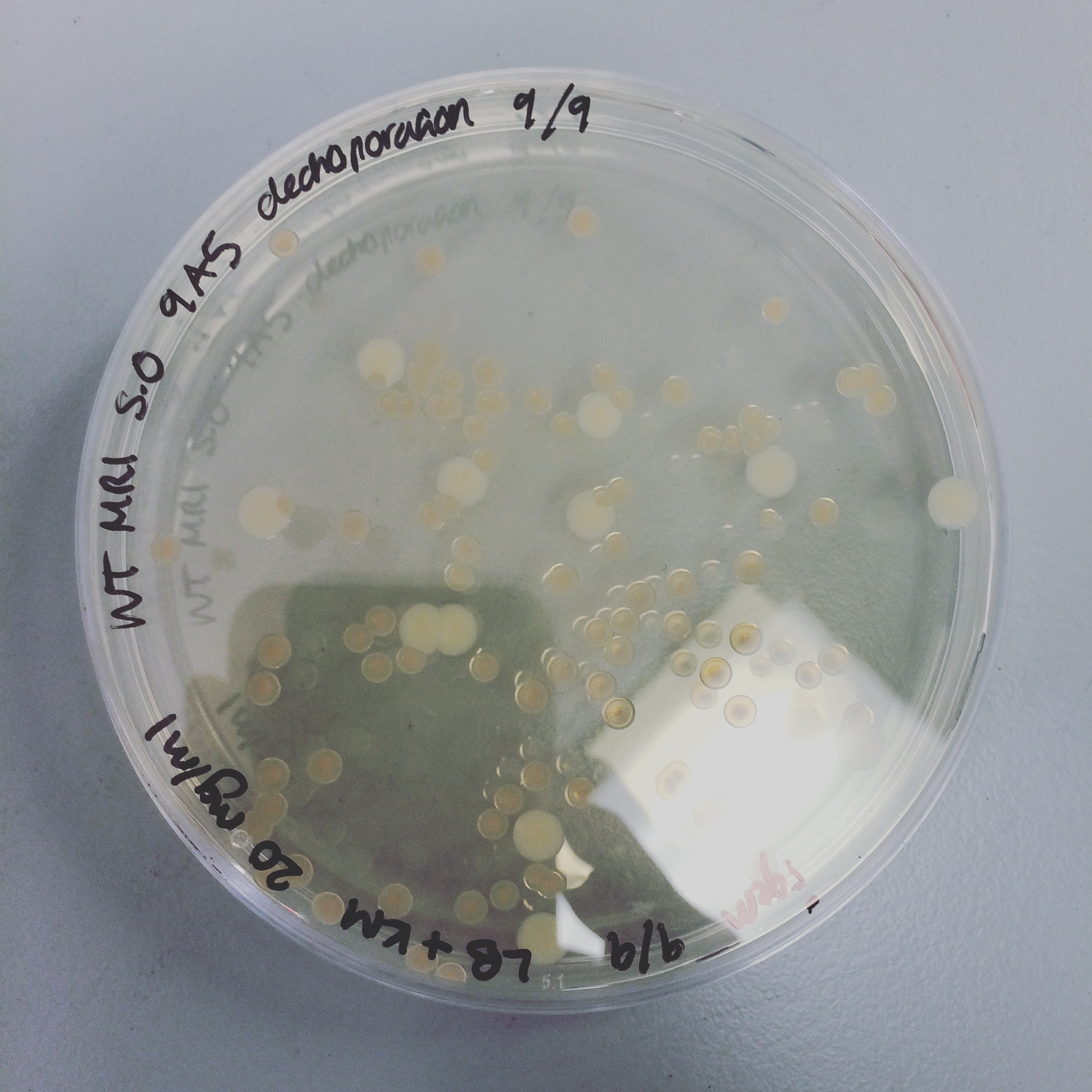
The triparental conjugation was continued from last week, however there were no S. oneidensis colonies containing the plasmid. Attempted electroporation to deliver the plasmid with HydABC into S. oneidensis. The first attempt did not yield any results, but the second attempt gave colonies of S.oneidensis MR-1 and ∆HydABC,HyaABCstrains S. oneidensis with both 8A5 and 9A5.
We grew∆HydABC,HyaABC and wildtype MR-1 strains for 24 hours in anaerobic conditions. We then used an electrochemical cell setup to analyse the activation of the hydrogenase enzymes, and the difference between the two strains. This would then serve as a control for later experiments.
Week 11 12.09-16.09
Attempted to start again with NiFe catalytic subunit cloning. PCR amplified with new primers and digested with a new source of EcoR1 and Pst1 as digests with our enzymes showed high amounts of smearing. Digested gel showed a band for HyaB N-terminus but not for HyaB C-terminus. Ligations, at ratios of 1:3, 1:5 and 1:7, were set up to be transformed the following week. Alongside HyaB N- and C-terminus, it was decided to excise HydABC out of the pBAD vector and ligate that into pSB1C3.
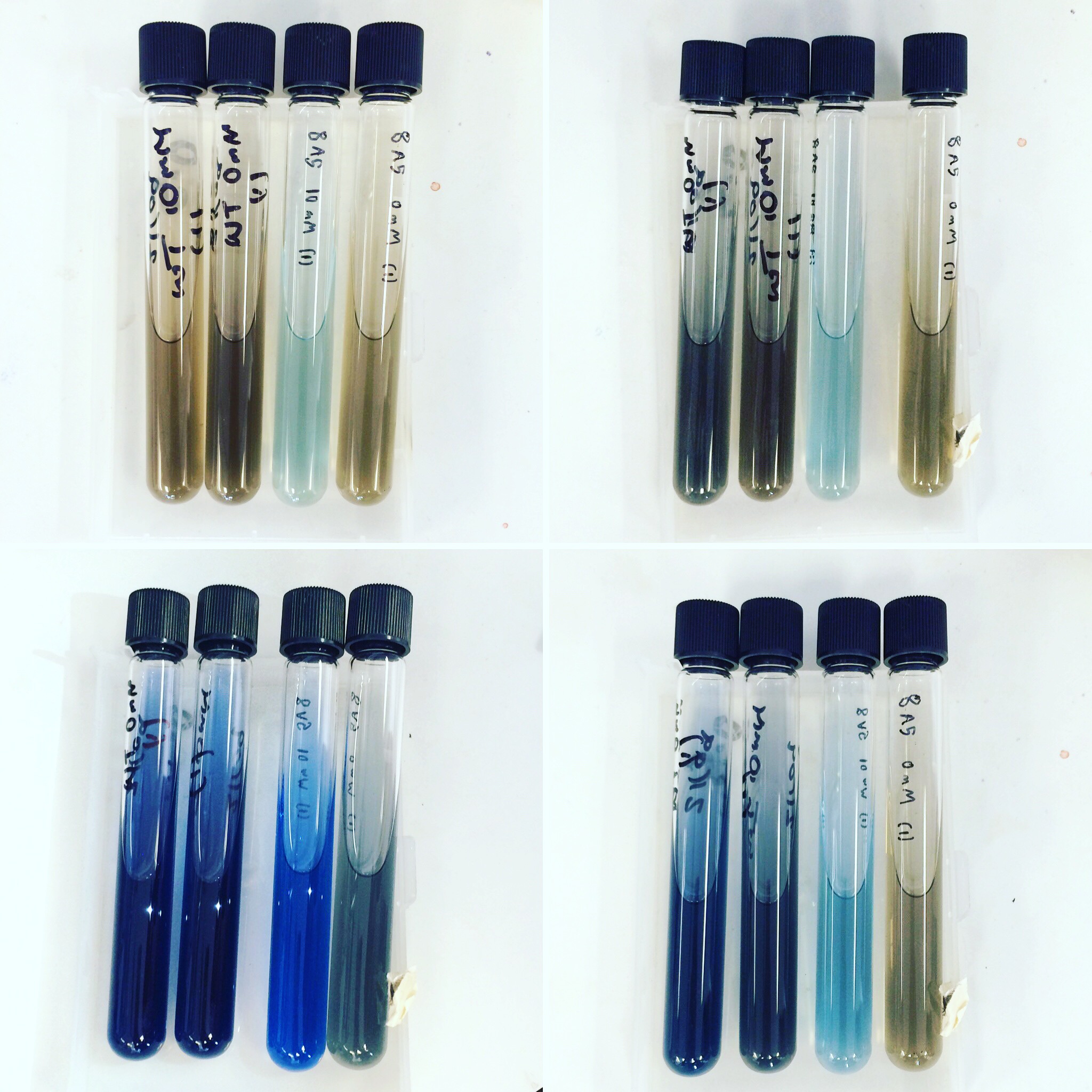
Mini-prepped colonies of the electroporation were sent off for sequencing. Results showed that only wild type S. oneidensis MR-1 has successfully taken up the HydABC C-terminally tagged i.e. 8A5, construct. Triparental conjugation and electroporation was continued with other strains but did not yield results. Protein expression trials (induced with 0mM, 1mM and 10mM arabinose) of the WT with 8A5, E.coli with 8A5 and 9A5 were performed using SDS-PAGE and western blot techniques.
Week 12 19.09-23.09
NiFe Hydrogenase subunits and HydABC were ligated and transformed into E.coli cells. All plates had colonies, however the digested gel samples showed that our insert was not in them this was most likely due to incomplete digestion of Bba_J04450. The DNA concentrations of HyaB C- and N-terminus were low. Thus, we decided to order new DNA, in order to re-attempt cloning of HyaB C- and N-terminus into pSB1C3
The triparental conjugations and electroporations from the previous week did not give any positive results. Protein expression trials (induced with 0mM, 1mM and 10mM arabinose) using SDS-PAGE and Western Blot were repeated for Mr1:HydABC and E.coli with 8A5 and 9A5.
Week 13 26.09-30.09
The digests of RFP were repeated to obtain a source of vector for new ligations for NiFe gene inserts and HydABC cluster. There were transformed into E.coli cells. All plates showed the ligations were successful as colonies had grown. The colonies were used to inoculate LB and mini prepped to perform a diagnostic colony PCR. Results showed correct ligations of all NiFe gene inserts, yielding more BioBricks.
Week 14 3.10-8.10
We grew wildtype MR-1 and MR-1 FeFe overexpression strains (our modified strain i.e. HydABC ligated into pBAD by Golden Gate cloning) in anaerobic conditions with varying concentrations of arabinose added. This showed the difference in expression between arabinose concentrations and also the effect of adding our FeFe HydABC hydrogenase cluster into the bacteria. This data could also be compared to MR1 without arabinose.