ReneVerhoef (Talk | contribs) |
ReneVerhoef (Talk | contribs) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 14: | Line 14: | ||
<div class="Panel"> | <div class="Panel"> | ||
<div class="Panel_Title"> | <div class="Panel_Title"> | ||
| − | + | Small molecule mediated scaffolds | |
</div> | </div> | ||
| Line 61: | Line 61: | ||
Read more | Read more | ||
</a> | </a> | ||
| − | <div class="Slide_Header" id="Project_title"> | + | <div class="Slide_Header" id="Project_title">T14-3-3 based scaffold proteins</div> |
<div class="Slide_Row"> | <div class="Slide_Row"> | ||
<div class="Column_50"> | <div class="Column_50"> | ||
| Line 108: | Line 108: | ||
<div class="Column_50"> | <div class="Column_50"> | ||
<p> | <p> | ||
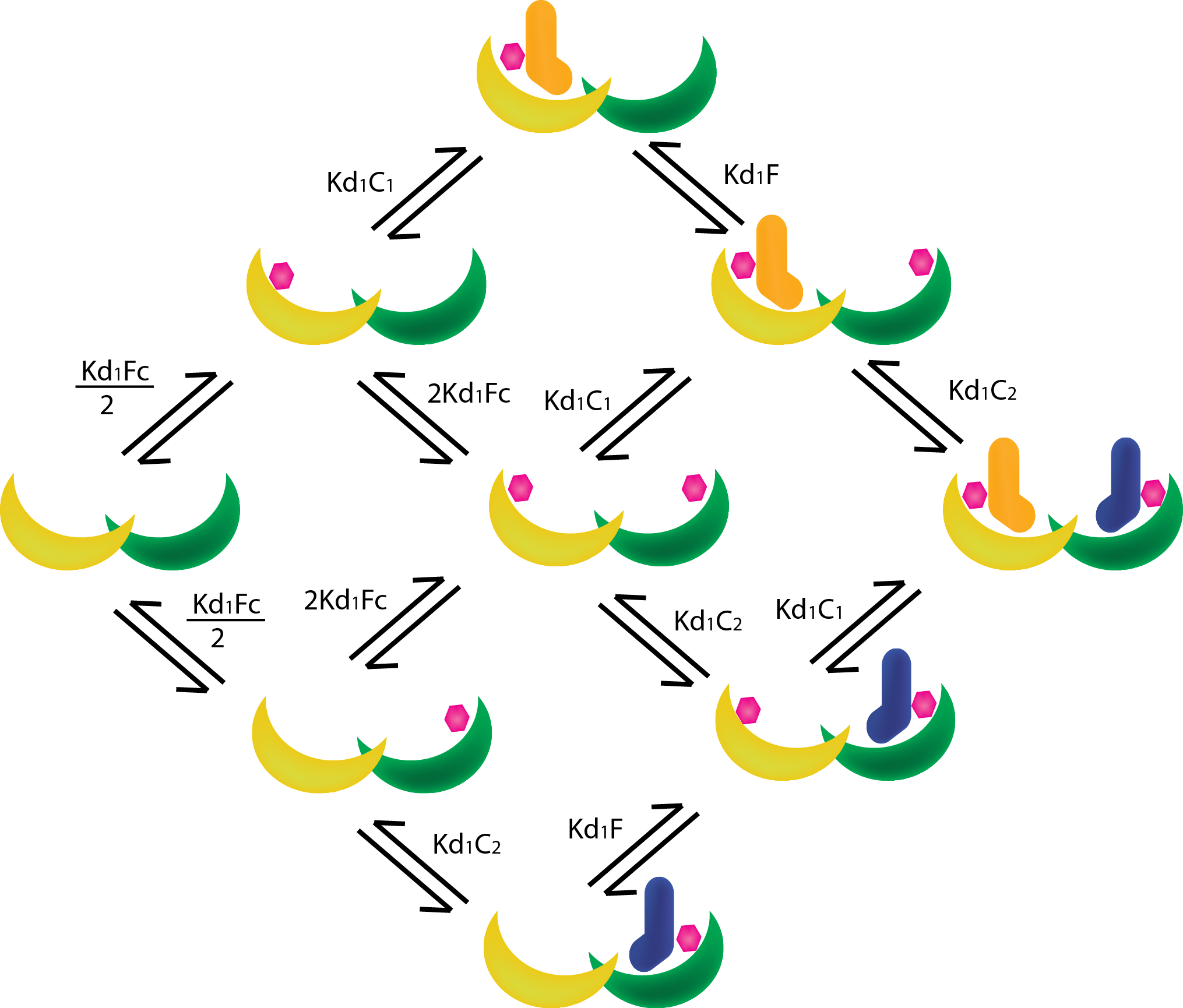
| − | In order to create | + | In order to create new heterodimeric scaffold it is essential to find suitable mutations to create a mutated T14-3-3/CT52 pair that is orthogonal to the wildtype. To find these mutations, The Rosetta software and a self-written protocol was used to determine the yet unknown properties of our newly designed pairs, a model based on Mass-Action and Michaelis-Menten kinetics was developed. |
</p> | </p> | ||
</div> | </div> | ||
| Line 134: | Line 134: | ||
</li> | </li> | ||
<li id="slide5"> | <li id="slide5"> | ||
| − | <a class="Button" href="https://2016.igem.org/Team:TU-Eindhoven/Results"> | + | <a class="Button" href="https://2016.igem.org/Team:TU-Eindhoven/Results/Introduction"> |
Read more | Read more | ||
</a> | </a> | ||
| Line 144: | Line 144: | ||
<div class="Column_50"> | <div class="Column_50"> | ||
<p> | <p> | ||
| − | The data we acquired in the lab | + | The performance of CT52 fused NanoBiT fragments was analysed by measuring the luminescence at varying concentrations. The data we acquired in the lab by NanoBit assays for our heterodimers was used in order to verify the quality or our mutations. For each newly found mutation set the functionality of the scaffold and the orthogonality with respect to the wildtype were determined. |
| + | Furthermore a caspase-9 assay was performed to test the activity of caspase-9 after increasing its concentration by T14-3-3. | ||
| + | |||
</p> | </p> | ||
</div> | </div> | ||
Latest revision as of 21:23, 19 October 2016

-
Read more
T14-3-3 based scaffold proteins
In the emerging field of synthetic biology, many new innovations arise. To use them as efficient and safe as possible, regulation is key. Therefore, iGEM TU Eindhoven is developing new kinds of scaffold proteins, based on 14-3-3 proteins. These scaffold proteins have a wide range of applications in nature and can be used to regulate systems in synthetic biology.
-
Read more
Team
The iGEM 2016 team of the Eindhoven University of Technology consists of 9 enthusiastic undergraduates of both Biomedical Engineering and Medical Sciences and Technology. Our members work very hard both inside and outside the lab, and have a great time working on our project and learn a lot thanks to iGEM.
-
Read more
Model
In order to create new heterodimeric scaffold it is essential to find suitable mutations to create a mutated T14-3-3/CT52 pair that is orthogonal to the wildtype. To find these mutations, The Rosetta software and a self-written protocol was used to determine the yet unknown properties of our newly designed pairs, a model based on Mass-Action and Michaelis-Menten kinetics was developed.
-
Read more
Human Practices
-
Read more
Results

The performance of CT52 fused NanoBiT fragments was analysed by measuring the luminescence at varying concentrations. The data we acquired in the lab by NanoBit assays for our heterodimers was used in order to verify the quality or our mutations. For each newly found mutation set the functionality of the scaffold and the orthogonality with respect to the wildtype were determined. Furthermore a caspase-9 assay was performed to test the activity of caspase-9 after increasing its concentration by T14-3-3.