(→Results) |
(→Results) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 12: | Line 12: | ||
[[File:T--Aix-Marseille--Toulouse_logo.jpg|link=https://2016.igem.org/Team:Toulouse_France|300px|350px|center]] | [[File:T--Aix-Marseille--Toulouse_logo.jpg|link=https://2016.igem.org/Team:Toulouse_France|300px|350px|center]] | ||
| − | + | ==Introduction== | |
We conceived a model in order to handle questions concerning the following situation: | We conceived a model in order to handle questions concerning the following situation: | ||
A bacterial growth is carried out in a bioreactor, continually supplied in substrate. | A bacterial growth is carried out in a bioreactor, continually supplied in substrate. | ||
| Line 24: | Line 24: | ||
As a plasmids can be a disadvantage for growth (energy spent into replicating processes) or a advantage (protection against a toxin) this question is hard to answer. But in this situation, where one type of plasmid can influence on the presence of the other type of plasmid in (and reciprocally) in the same bacteria, the question become too tough to answer and only a mathematical model can resolve such a interrogation! | As a plasmids can be a disadvantage for growth (energy spent into replicating processes) or a advantage (protection against a toxin) this question is hard to answer. But in this situation, where one type of plasmid can influence on the presence of the other type of plasmid in (and reciprocally) in the same bacteria, the question become too tough to answer and only a mathematical model can resolve such a interrogation! | ||
| − | + | ==Equations== | |
| − | + | ===Development of the model=== | |
In 1967 Fredrickson et al. <ref name="Fredrickson1967">[https://pdfs.semanticscholar.org/1873/0fa936b17078cfe2b0ab1f74d44eae002758.pdf Fredrickson AG, Ramkrishna D, Tsuchiya H (1967) Statistics and dynamics of correct pro- caryotic cell populations. Mathematical Biosciences 1: 327–374.]</ref> studied mathematically | In 1967 Fredrickson et al. <ref name="Fredrickson1967">[https://pdfs.semanticscholar.org/1873/0fa936b17078cfe2b0ab1f74d44eae002758.pdf Fredrickson AG, Ramkrishna D, Tsuchiya H (1967) Statistics and dynamics of correct pro- caryotic cell populations. Mathematical Biosciences 1: 327–374.]</ref> studied mathematically | ||
| Line 245: | Line 245: | ||
</nowiki> | </nowiki> | ||
| − | + | ===Objectives=== | |
The aim of studying the behaviour of this model is to investigate how | The aim of studying the behaviour of this model is to investigate how | ||
| Line 253: | Line 253: | ||
the culture progresses, and how this depends on the various parameters in the model. | the culture progresses, and how this depends on the various parameters in the model. | ||
| − | + | ===Proposition: Adding contention=== | |
We need to introduce a modification to equation 3 (the state dependant | We need to introduce a modification to equation 3 (the state dependant | ||
| Line 299: | Line 299: | ||
gene copy and without anti-toxin to the $IC_{50}$. | gene copy and without anti-toxin to the $IC_{50}$. | ||
| − | + | ==Program== | |
We made a program to simulate the model corresponding to these equations, using a bayesian algorithm. | We made a program to simulate the model corresponding to these equations, using a bayesian algorithm. | ||
| − | + | It is programmed in C to maximize performance, using the GNU scientific library. | |
Plots were made using R with ggplot2. | Plots were made using R with ggplot2. | ||
| Line 309: | Line 309: | ||
[[https://static.igem.org/mediawiki/2016/f/f8/T--Aix-Marseille--program-toulouse.zip download]] | [[https://static.igem.org/mediawiki/2016/f/f8/T--Aix-Marseille--program-toulouse.zip download]] | ||
| − | + | ==Results== | |
[[File:T--Aix-Marseille--model-plot-d20-70.png|300px|350px|left|thumb]] | [[File:T--Aix-Marseille--model-plot-d20-70.png|300px|350px|left|thumb]] | ||
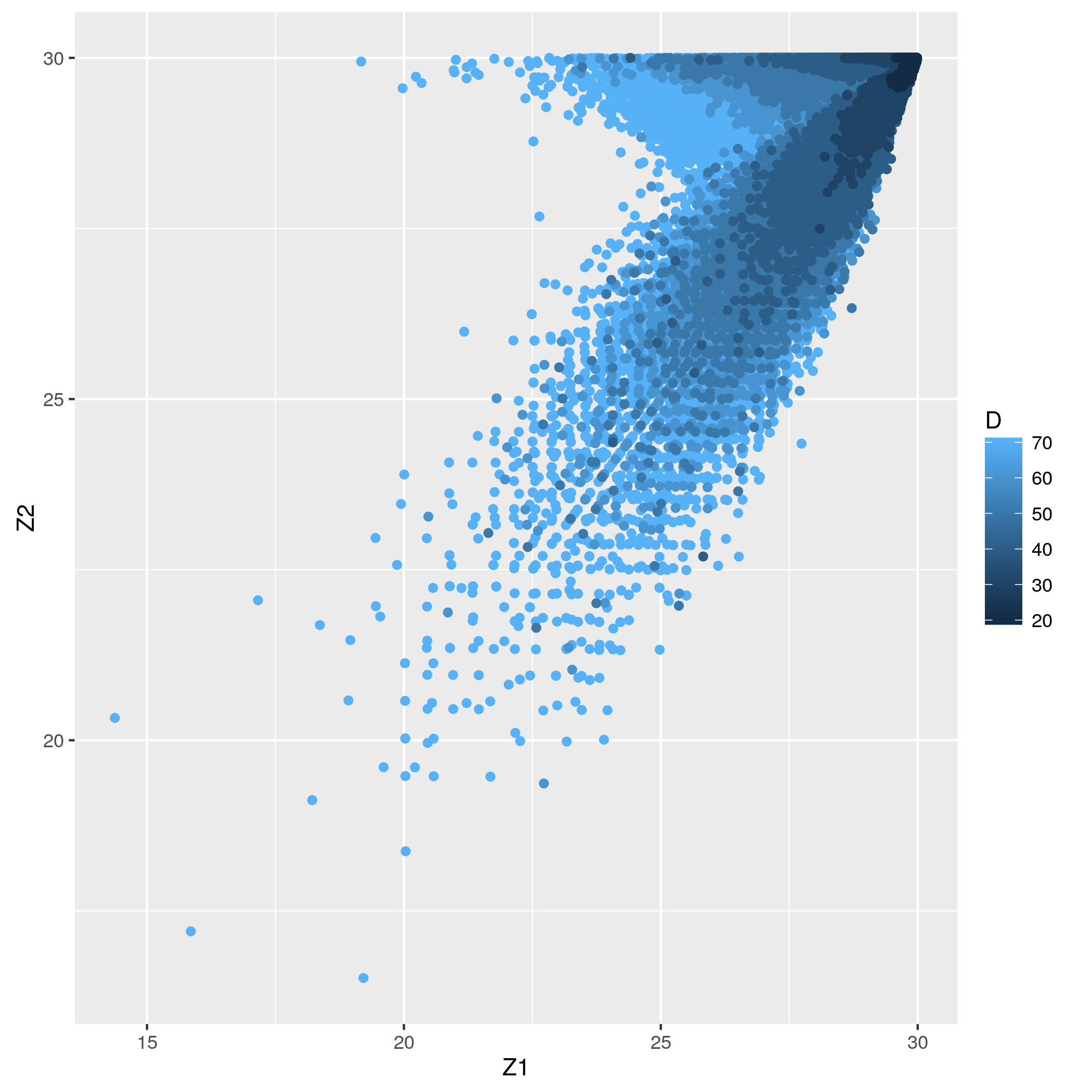
| − | We can see that with D | + | We can see that with D increasing, corresponding to more bacteria growing, more bacteria tend to lose their plasmids and to have various numbers of plasmid. |
| − | + | ||
| + | These results can be interpreted by the fact that if D is increasing, thus $µ$ also increases, then bacteria grow faster, and divide faster. | ||
| + | With small values of D, bacteria grow slowly and all plasmids have time to be replicated between each cell division. | ||
| + | That is why for these values, most bacteria still have maximum values of plasmids and are all in the same situation hence the focusing of the plot for small values of D. | ||
| + | If D is higher time for each bacteria to replicate their plasmids between each division is reduced, and the situation of each individual bacteria becomes different, explaining the scattering on the plot. | ||
| + | This show that one of the main factor influencing the loss of plasmid is the growth of bacteria. Of course other factor have shown impact as replication rate of plasmids and the relative toxicity of each toxin. | ||
| + | |||
| + | <div style="clear:both"></div> | ||
| + | |||
| + | ==Bibliography== | ||
| − | |||
<references/> | <references/> | ||
| + | |||
| + | {{:Team:Aix-Marseille/Template-Footer}} | ||
Model
Introduction
We conceived a model in order to handle questions concerning the following situation: A bacterial growth is carried out in a bioreactor, continually supplied in substrate. Bacteria can possess 2 type of plasmids:
- plasmids 1 carry the toxin A gene and the anti-toxin B gene
- plasmids 2 carry the toxin B gene and the anti-toxin A gene
So during the growth each bacterium can have no plasmids, either one type of plasmids, or both types.
The aim of the model is to assess the evolution of plasmids throughout the culture, to determine which parameters can matter in the loss of those plasmids, and to precise what are the probabilities for a bacteria to loose its plasmids during cell division. As a plasmids can be a disadvantage for growth (energy spent into replicating processes) or a advantage (protection against a toxin) this question is hard to answer. But in this situation, where one type of plasmid can influence on the presence of the other type of plasmid in (and reciprocally) in the same bacteria, the question become too tough to answer and only a mathematical model can resolve such a interrogation!
Equations
Development of the model
In 1967 Fredrickson et al. [1] studied mathematically development of a bacterial population, under the assumptions of a large population of independant bacteria in a well mixed solution of constant volume. The large population ensures that for the population the expectation value is a good estimate of the average. The bacteria being independant ensures that the behaviour of each individual depends only on its internal state $\mathbf{z}$ and the conditions $\mathbf{c}$ which are the same for all individuals. The volume is well mixed so the conditions $\mathbf{c}$ which are the same everywhere. The volume is constant so that the population caracteristics can be evaulated by integration over the volume. In their development $\mathbf{z}$ and $\mathbf{c}$ are considered to be arbitrary vector quantities.
From this starting point they develop a pair of *master equations of change* to describe the evolution of the population:
\begin{equation} \begin{gathered} \frac{\partial}{\partial t} W_{\mathbf{Z}}(\mathbf{z},t) + \nabla_{\mathbf{Z}} \cdotp [(\mathbf{\beta} \cdotp \bar{\mathbf{R}}(\mathbf{z,c}) W_{\mathbf{Z}}(\mathbf{z},t)] \\ = 2 \int \sigma (\mathbf{z',c}) p(\mathbf{z,z',c}) W_{\mathbf{Z}}(\mathbf{z'},t)\mathrm{d}v' - (D+\sigma (\mathbf{z,c})) W_{\mathbf{Z}}(\mathbf{z},t)\end{gathered} \end{equation} \begin{equation} \frac{d\mathbf{c}}{dt} = D(\mathbf{c_f} - \mathbf{c} ) + \mathbf{\gamma} \cdotp \int \bar{\mathbf{R}}(\mathbf{z,c}) W_{\mathbf{Z}}(\mathbf{z},t)\mathrm{d}v \end{equation}
In these equations the various symbols are as follows:
| $\mathbf{z}$ | Vector for internal state of a bacteria. |
| $\mathbf{c}$ | Time dependant vector for conditions. |
| $W_\mathbf{z} (\mathbf{z},t)$ | Distribution of bacteria in z, t space. |
| $\overline{\mathbf{R}}(\mathbf{z},c)$ | The expected value or the reaction rate vector in z, t space. |
| $\sigma (\mathbf{z,c})$ | Rate of fision for bacteria in z, c space. |
| $p(\mathbf{z,z',c}))$ | Partitioning probability of generating a child in state z from a parent in state z'. |
| $\nabla_\mathbf{z}\cdot\mathbf{V}$ | $\sum \frac{\partial}{\partial z_i}\mathbf{V}_i$ |
| $\mathrm{d}v'$ | Integral over state space v' |
| $D$ | Dilution rate of the culture (for femrenters). |
| $\beta$ | Stochiometric matrix for cellular substances. |
| $\gamma$ | Stochiometric matrix for extra-cellular substances. |
| $\bar{\dot{\mathbf{V}}}(\mathbf{z,c}) = \mathbf{\beta} \cdotp \bar{\mathbf{R}}(\mathbf{z,c}) $ | The expected internal state change rate vector. |
| $ -\mathbf{\gamma} \cdotp \bar{\mathbf{R}}(\mathbf{z,c}) $ | The expected consumation of substances in the environment by a cell in state $\mathbf{z}$ . |
Thus for a particular problem in hand it is necessary to chose
$\mathbf{z}$ and $\mathbf{c}$ that represent the state of cells and the
media. Then the matrices and functions $\beta$, $\gamma$,
$\bar{\mathbf{R}}(\mathbf{z,c})$, $\sigma (\mathbf{z',c})$ and
$p(\mathbf{z,z',c}) $ need to be defined for the problem considered.
Finally the inital conditions $W_{\mathbf{Z}}(\mathbf{z},t)$ and
$\mathbf{c}_0 $ and growth conditions $D$ and $\mathbf{c}_f $ need to be
fixed.
For the problem in hand, plasmid maintenance during growth with 2 different plasmids, and attempting to find a simple solution to the problem we propose a 3 variable internal state vector:
$$\mathbf{z} = \begin{bmatrix} z_0 \\ z_1 \\ z_2 \end{bmatrix} = \begin{bmatrix}\textrm{Cell maturity} \\ \textrm{count of plasmid 1} \\ \textrm{count of plasmid 2} \end{bmatrix}$$
In this internal state vector: $z_0$ is a mesure of the growth of the bacteria, encompassing such things as size, number of chromosomes and mass; $z_1$ and $z_2$ represent the number of copies of each plasmid. For the external conditions we propose simply the substrate concentration $S$. The maturity parameter has a minimum value of 1 and must increase to 2 before division can occur.
For the rates of change of the internal state vector then we propose for the bacterial maturity to extend the development presented in Shene et al. [2] to include 2 plasmids and incorporate the cell maturity as a state variable. This gives:
\begin{equation} \dot{z}_0 (\mathbf{z},S) = \mu = \mu _{max} \frac{S}{K_S+S} \frac{K_{z_1}}{K_{z_1}+z_1^{m_1}} \frac{K_{z_2}}{K_{z_2}+z_2^{m_2}} \end{equation}
Here $ \mu _{max}$ is the maximum growth rate $hr^{-1}$: $\mu (\mathbf{z,S})$ the growth rate ; $K_S$ is the Monod constant in $g/l$ for the substrate; $K_{z_1}$ is the inhibition constant for plamid number 1 in (plasmids per cell)$^{m_1}$, and $m_1$ the Hill coefficient for the cooperativity of inhibition. $K_{z_2}$ and $m_2$ represent the same parameters for plasmid 2.
For plasmid replication rate we propose, again following Shene et al. [2], the empirical relationship :
\begin{equation} \dot{z}_1 (\mathbf{z},S) = \begin{cases} k_1 \frac{\mu (\mathbf{z},S)}{K_1 + \mu (\mathbf{z},S)} ( z_{1_{max}} - z_1 ) & \text{if } z_1 \geq 1.0 \\ 0 & \text{if } 0.0 \leq z_1 < 1.0 \\ \end{cases} \end{equation}
This relation, and equivalent one for plasmid number 2 $\dot{z}_2 (\mathbf{z,S})$ is designed to satisfy the boundary conditions of no reproduction if there is less than 1 plasmid in the cell, and a maximum copy number of $z_{1_{max}}$. This introduces the parameters $k_1$ and $K_1$ which are respectively the plasmid replication rate (in $hr^{-1}$) and the inhibition constant (also in $hr^{-1}$). The inhibition constant reduces plasmid replication rate at slower growth rates.
Notice that here we have directly defined $\bar{\dot{\mathbf{V}}}(\mathbf{z,c})$ rather than $\beta $ and $\bar{\mathbf{R}}(\mathbf{z,c})$. For the growth yield we propose :
\begin{equation} \gamma \cdotp \bar{\mathbf{R}}(\mathbf{z,c}) = \alpha \mu(\mathbf{z},S) \end{equation}
The remaining functions and parameters in equations 1 and 2 are the division rate $\sigma (\mathbf{z,c})$ and the partitioning function $p(\mathbf{z,z',c}) $. There is less consensus in the litterature for an at least empirically appropriate form for these equations. To remain simple we propose:
\begin{equation} \sigma (\mathbf{z,c}) = \sigma \times H[2.0] = \begin{cases} 0 & \text{if } z_0 < 2.0 \\ \sigma & \text{if } z_0 \geq 2.0 \end{cases} \end{equation}
Here we assume that there is a fixed rate of division $\sigma $ once cells are big enough to divide ($H[]$ is the Heaviside function).
\begin{equation} p(\mathbf{z,z',c}) = p(z_0,z'_0) \times p(z_1,z'_1) \times p(z_2,z'_2) \end{equation} \begin{equation} p(z_0,z'_0) = \delta_{z_0,\frac{z'_0}{2}} = \begin{cases} 1 & \text{if } z_0 = z'_0/2.0 \\ 0 & \text{if } z_0 \ne z'_0/2.0. \end{cases} \end{equation} \begin{equation} p(z_1,z'_1) = 0.5^{z'_1} \begin{pmatrix} z_1 \\ z'_1 \\ \end{pmatrix} = 0.5^{z'_1} \frac{z'_1 !}{(z'_1-z_1)! z_1 !} \end{equation}
In these equations we assume that the partitioning of the three internal state variables are independant. That cells divide exactly in half, that is the maturity parameter is exactly halved when the cells divide ($\delta$ is a Kronecker delta function). That the two plasmids segregate independantly and as individual plasmids according to a binomial distribution. These assumptions are probably the most suspect in the model.
This initial version of the model has no contention, that is $z_1$ and $z_2$ do not influence the growth rate $\mu $. In order to develop the model for the system envisaged this needs to be introduced.
Substituting into the equations 1 and 2 we obtain:
\begin{multline} \frac{\partial}{\partial t} W_{\mathbf{Z}}(\mathbf{z},t) + [\nabla_{\mathbf{Z}} \cdotp \bar{\dot{\mathbf{Z}}}(\mathbf{z},S)] W_{\mathbf{Z}}(\mathbf{z},t) + \sum_i \bar{\dot{z_i}} \times \frac{\partial}{\partial z_i} W_{\mathbf{Z}}(\mathbf{z},t) \\ = 2 \sigma \int_{z'_0>2.0} \delta_{z_0,\frac{z'_0}{2}} \times 0.5^{z'_1+z'_2} \times \begin{pmatrix} z_1 \\ z'_1 \\ \end{pmatrix} \times \begin{pmatrix} z_2 \\ z'_2 \\ \end{pmatrix} \times W_{\mathbf{Z}}(\mathbf{z'},t)\mathrm{d}v' \\ - (D+\sigma H[2.0] W_{\mathbf{Z}}(\mathbf{z},t)) \end{multline} \begin{equation} \frac{d\mathbf{c}}{dt} = D(c_f - c ) - \alpha \int \mu (\mathbf{z},S) W_{\mathbf{Z}}(\mathbf{z},t)\mathrm{d}v \end{equation}
Objectives
The aim of studying the behaviour of this model is to investigate how growth conditions $D, S_f$ and time modulate the development of the population in state space $W_{\mathbf{Z}}(\mathbf{z},t)$. In particular we are interested in finding how the number of bacteria without plasmids $W_{\mathbf{Z}}([z_0,0,0],t)$ in the culture progresses, and how this depends on the various parameters in the model.
Proposition: Adding contention
We need to introduce a modification to equation 3 (the state dependant growth rate) in which the equilibrium between $z_1$ and $z_2$ features. This modification is to take into account that each toxin should be inhibited by anti-toxin for maximal growth. A possibility is that we consider that for each toxin anti-toxin pair:
- the genes produce toxin at a constant rate dependant on the numberof copies $k_1\times z_1$,
- the anti-toxin genes produce anti-toxin at a rate dependant on the number of copies $k_2\times z_2$,
- anti-toxin instantly and irreversible kills the toxin in a stochiometric manner.
- undestroyed toxin disappears at a constant rate, by dilution and other pathways $k_3$,
- the concentration of toxin is at a dynamic steady state, i.e. the rates of production and disappearance are equal.
- growth in inhibited in an exponential manner by free toxin with a characteristic $IC_{50}$.
This model of toxin anti-toxin interaction takes into account the bacteriostatic nature of most such toxins, and the presence of measurable $IC_{50}$ values. Clearly a more realsitic model would need to take into account cell volume, protein synthesis rates etc. Nevertheless, this simple model gives: $$ \begin{align} \mu &= \mu_{0} \times e^{-\frac{[Toxin]}{IC_{50}}} \\ [Toxin] &= max(0,\frac{k_1z_1-k_2z_2}{k_3}) \\ \mu &= \mu_{0} \times min(1.0,e^{-k_a(z_1-k_bz_2)}) \\ k_a &= \frac{k_1}{k_3 \times IC_{50}} \\ k_b &= \frac{k_2}{k_1} \end{align} $$
Incorporating 2 toxin anti toxin pairs, if we assume that the production rate ratio is the same for both, $k_b$, is independant of the system we have:
\begin{equation} \mu = \mu_{0} \times min(1,e^{-k_a(z_1-k_bz_2)})) \times min(1,e^{-k_a(z_2-k_bz_1)})) \end{equation}
where $\mu_0$ is given by equation 3. For each toxin the efficiency parameter is a measure of ratio of toxin accumulation in cells with one gene copy and without anti-toxin to the $IC_{50}$.
Program
We made a program to simulate the model corresponding to these equations, using a bayesian algorithm.
It is programmed in C to maximize performance, using the GNU scientific library.
Plots were made using R with ggplot2.
[download]
Results
We can see that with D increasing, corresponding to more bacteria growing, more bacteria tend to lose their plasmids and to have various numbers of plasmid.
These results can be interpreted by the fact that if D is increasing, thus $µ$ also increases, then bacteria grow faster, and divide faster. With small values of D, bacteria grow slowly and all plasmids have time to be replicated between each cell division. That is why for these values, most bacteria still have maximum values of plasmids and are all in the same situation hence the focusing of the plot for small values of D. If D is higher time for each bacteria to replicate their plasmids between each division is reduced, and the situation of each individual bacteria becomes different, explaining the scattering on the plot. This show that one of the main factor influencing the loss of plasmid is the growth of bacteria. Of course other factor have shown impact as replication rate of plasmids and the relative toxicity of each toxin.