(→InterLab) |
|||
| (38 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{LMU-TUM_Munich|navClass= | + | {{LMU-TUM_Munich|navClass=interlab}} |
__notoc__ | __notoc__ | ||
| − | |||
| − | + | =InterLab= | |
| + | <div class="white-box"> | ||
| + | For the purpose of the third interlaboratory measurement study organized by iGEM about the reproducibility of fluorescence measurements, five different GFP expression plasmids were transformed and expressed in the growth rate of ''E. coli'' bacteria. The corresponding expression levels of the different constructs were determined by measuring OD<sub>600</sub> and fluorescence by a plate reader over time. | ||
| + | </div> | ||
==Introduction== | ==Introduction== | ||
| + | <div class="white-box"> | ||
| + | Science is only as good as the experiments underlying it - and experiments are only as good as the accuracy of their measurements. Once again, iGEM has called labs all around the world to their instruments, in order to determine if a seemingly same experiment really is the same experiment everywhere - or whether the differences in instrument architecture and accuracy may make reproducibility a problem. For the third iGEM interlab study, fluorescence measurements with five different GFP expression plasmids in E. coli were performed. The growth rate of the bacteria and the corresponding expression levels of the different GFP constructs were determined via a plate reader. We are glad to be able to do our part for this crucial issue, especially since the competition is such a great place to gather data from all around the world! | ||
| + | </div> | ||
| − | + | ==Materials & Methods== | |
| + | <div class="white-box"> | ||
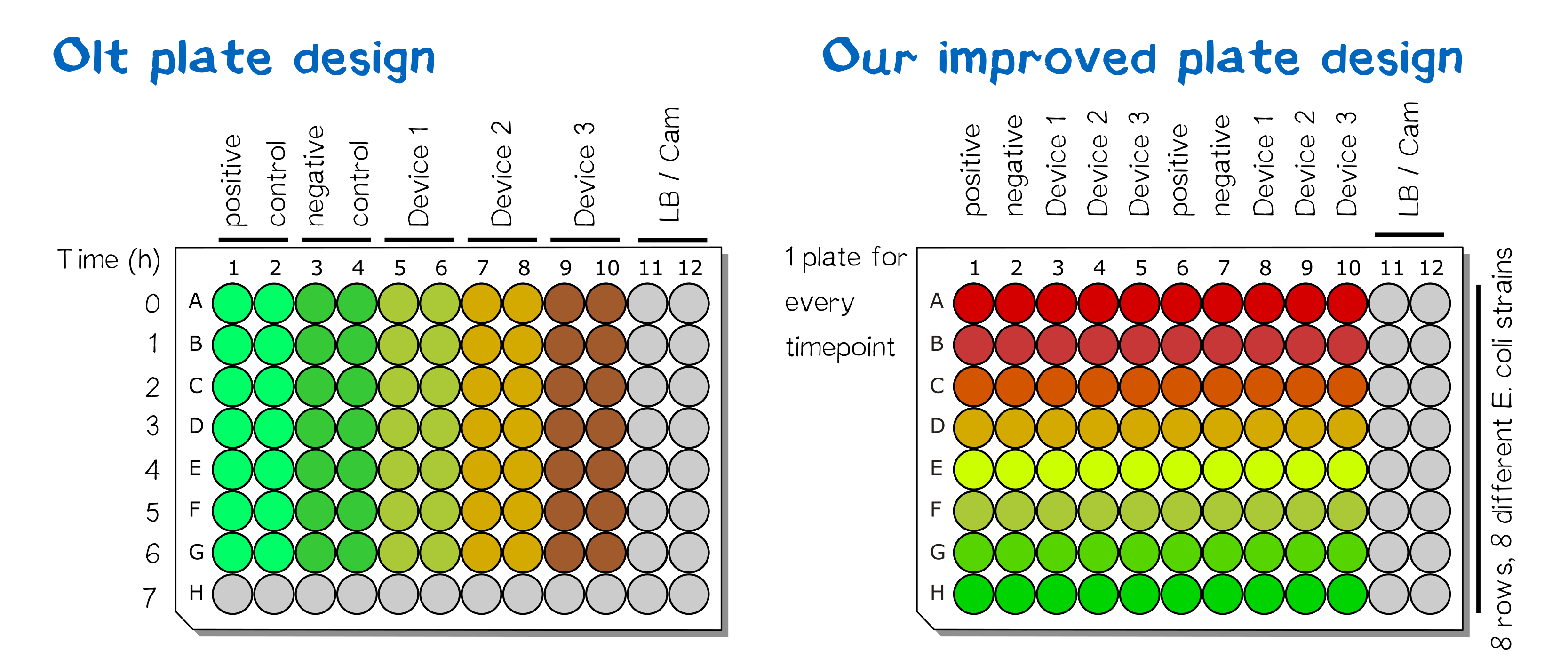
[[File:Muc16_Interlab_masterblock.jpg|thumb|right|250px| <i><b>Figure 1:</b></i> Masterblock layout. For details see figure 3.]] | [[File:Muc16_Interlab_masterblock.jpg|thumb|right|250px| <i><b>Figure 1:</b></i> Masterblock layout. For details see figure 3.]] | ||
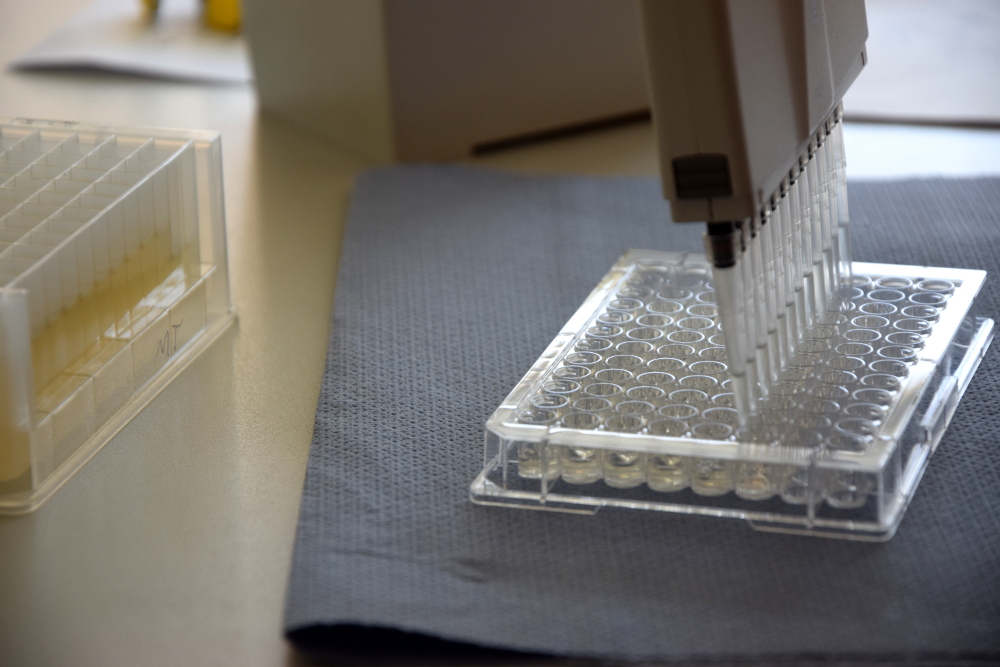
[[File:Muc16_Interlab_Microtest_Plate.jpg|thumb|right|250px| <i><b>Figure 2:</b></i> Transferring samples into microtest plate.]] | [[File:Muc16_Interlab_Microtest_Plate.jpg|thumb|right|250px| <i><b>Figure 2:</b></i> Transferring samples into microtest plate.]] | ||
| − | + | [[File:Muc16_Interlabplatedesign_001.png|thumb|right|250px| <i><b>Figure 3:</b></i> Standard layout and alternative layout for multi strain measurement of 96 well plate.]] | |
| − | + | ||
| − | [[File: | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
===Materials=== | ===Materials=== | ||
*[http://parts.igem.org/Help:InterLab_Measurement_Kit InterLab Measurement Kit] | *[http://parts.igem.org/Help:InterLab_Measurement_Kit InterLab Measurement Kit] | ||
| Line 30: | Line 23: | ||
*Masterblock: [https://shop.gbo.com/en/row/articles/catalogue/article/5_0060_0020_0020_0050/14284/ MASTERBLOCK®, 96 WELL, 2 ML, PP, V-BOTTOM, NATURAL, SINGLE PACKED; greiner bio-one; item number: 780270 ] | *Masterblock: [https://shop.gbo.com/en/row/articles/catalogue/article/5_0060_0020_0020_0050/14284/ MASTERBLOCK®, 96 WELL, 2 ML, PP, V-BOTTOM, NATURAL, SINGLE PACKED; greiner bio-one; item number: 780270 ] | ||
*''E. coli'' strains: ''DH5α'', ''W3110'', ''KS272'', ''XL-1 blue'', ''JM83'', ''OriB'', ''10β NEB turbo'' | *''E. coli'' strains: ''DH5α'', ''W3110'', ''KS272'', ''XL-1 blue'', ''JM83'', ''OriB'', ''10β NEB turbo'' | ||
| − | |||
===Methods=== | ===Methods=== | ||
| − | |||
====Given protocol==== | ====Given protocol==== | ||
| − | |||
*According to provided [https://static.igem.org/mediawiki/2016/c/c5/InterLab_iGEM2016_Plate_Reader_Protocol_Updated_July.pdf plate reader protocol] | *According to provided [https://static.igem.org/mediawiki/2016/c/c5/InterLab_iGEM2016_Plate_Reader_Protocol_Updated_July.pdf plate reader protocol] | ||
| − | |||
====Multi-measurement protocol==== | ====Multi-measurement protocol==== | ||
| − | |||
All five plasmids were transformed in different E. coli strains. Single colonies were picked and grown at 220 rpm and 37 °C in 1 mL LB liquid culture per well in one Masterblock over night. The layout of the plate can be found in figure 1 and figure 3. On the next day OD<sub>600</sub> were measured by pipetting 100 µL of each well into a microtest plate with the same layout using a 12 channel pipette. According to the results new diluted LB liquid cultures were mixed in a new Masterblock with the same layout and incubated at 37 °C for 6 hours at 220 rpm. At the beginning and every hour a sample was taken by transferring 100 µL of each well into a new microtest plate with the same layout, see figure 2. | All five plasmids were transformed in different E. coli strains. Single colonies were picked and grown at 220 rpm and 37 °C in 1 mL LB liquid culture per well in one Masterblock over night. The layout of the plate can be found in figure 1 and figure 3. On the next day OD<sub>600</sub> were measured by pipetting 100 µL of each well into a microtest plate with the same layout using a 12 channel pipette. According to the results new diluted LB liquid cultures were mixed in a new Masterblock with the same layout and incubated at 37 °C for 6 hours at 220 rpm. At the beginning and every hour a sample was taken by transferring 100 µL of each well into a new microtest plate with the same layout, see figure 2. | ||
| − | + | </div> | |
| − | + | ||
==Results== | ==Results== | ||
| + | <div class="white-box"> | ||
| + | Samples were prepared according to our multi-measurement protocol and the standard protocol. The results for DH5α <i>E. coli</i> strain are plotted in figure 4-8. | ||
| − | + | <gallery mode="packed-hover"> | |
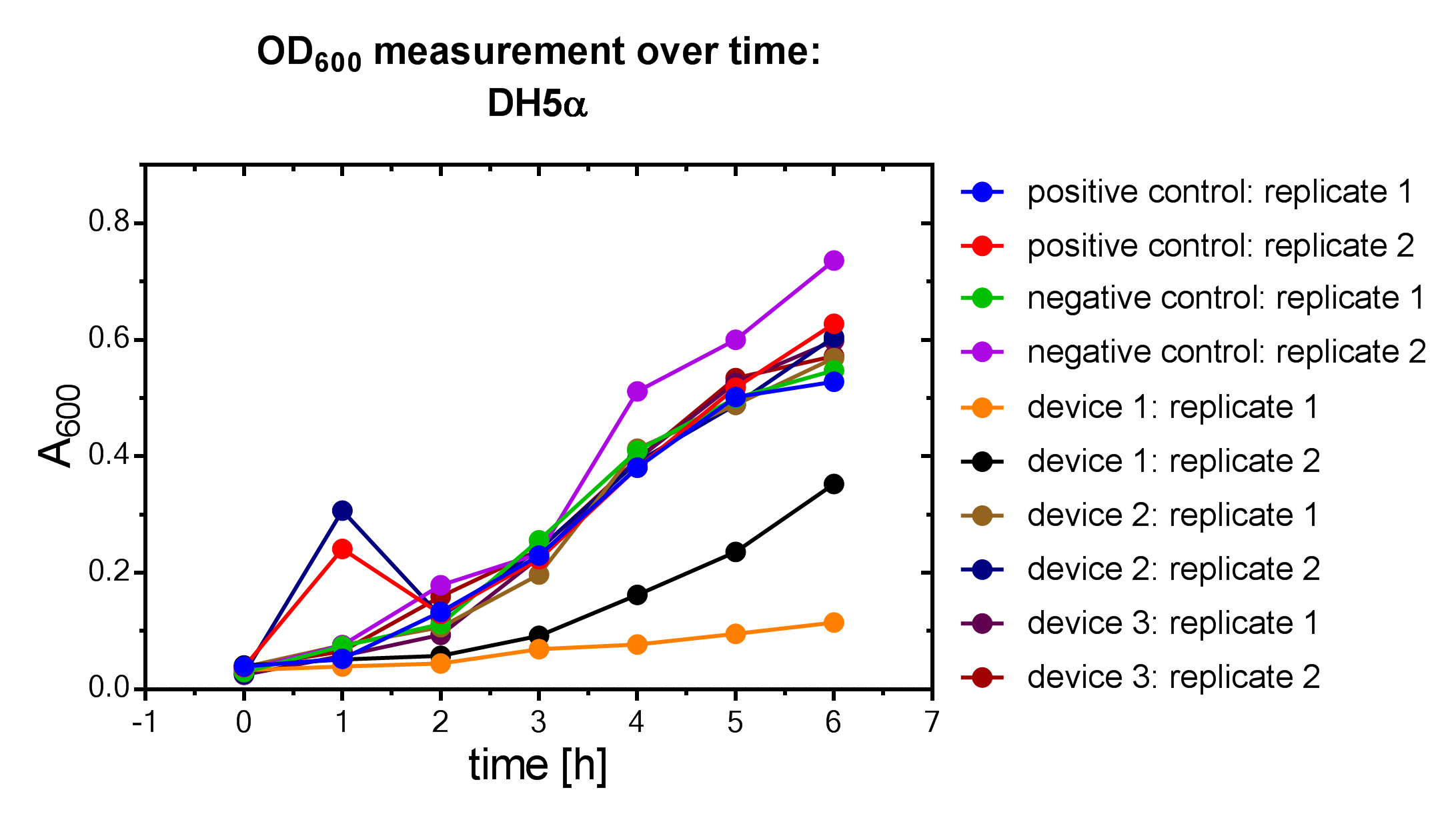
| + | File:Muc16_Interlab_od_protocol.png| <i><b>Figure 4:</b></i> OD<sub>600</sub> measurement of DH5α <i>E. coli </i> strain. | ||
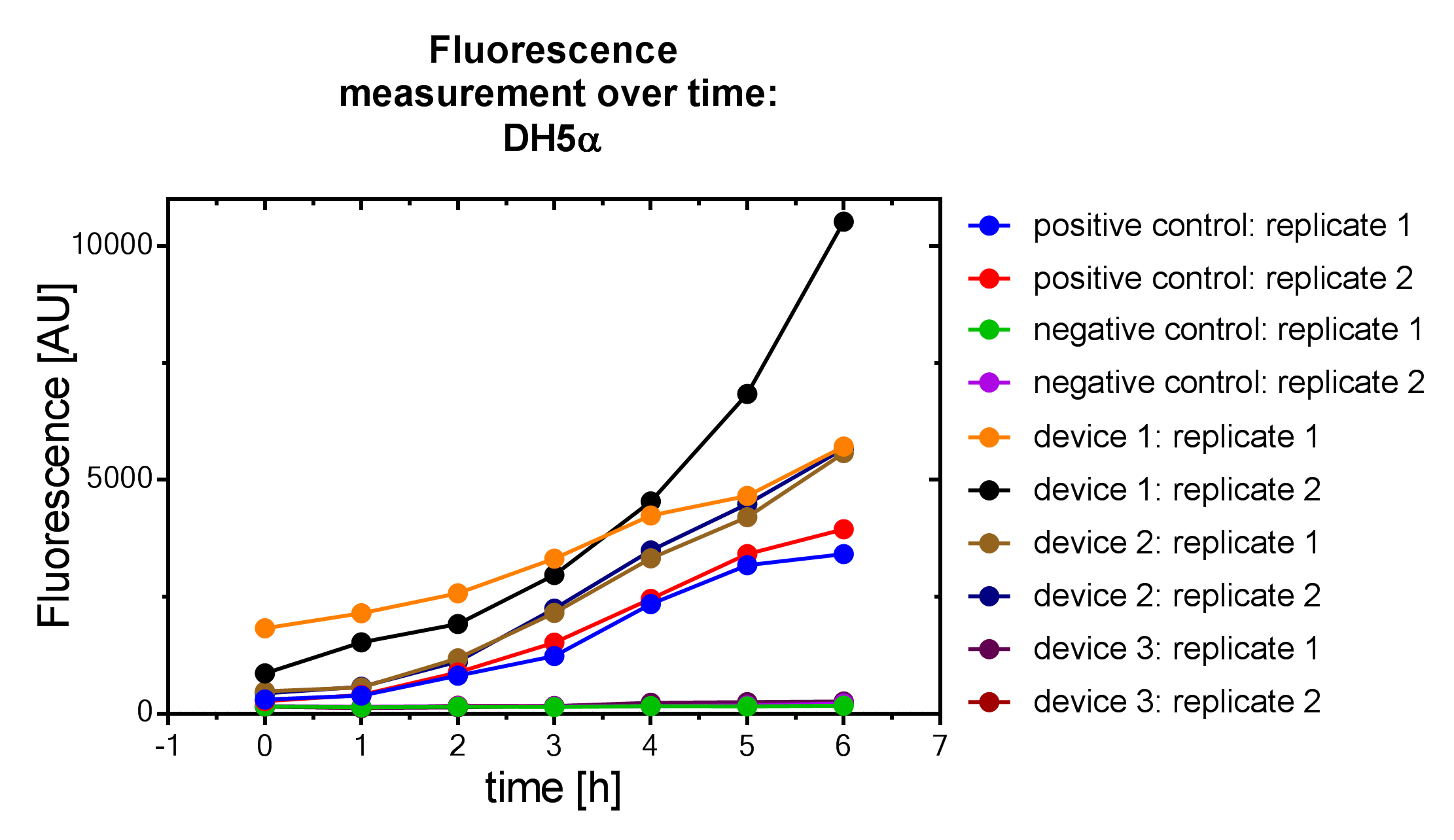
| + | File:Muc16_Interlab_fl_protocol.png| <i><b>Figure 5:</b></i> Fluorescence measurement of DH5α <i>E. coli </i> strain. | ||
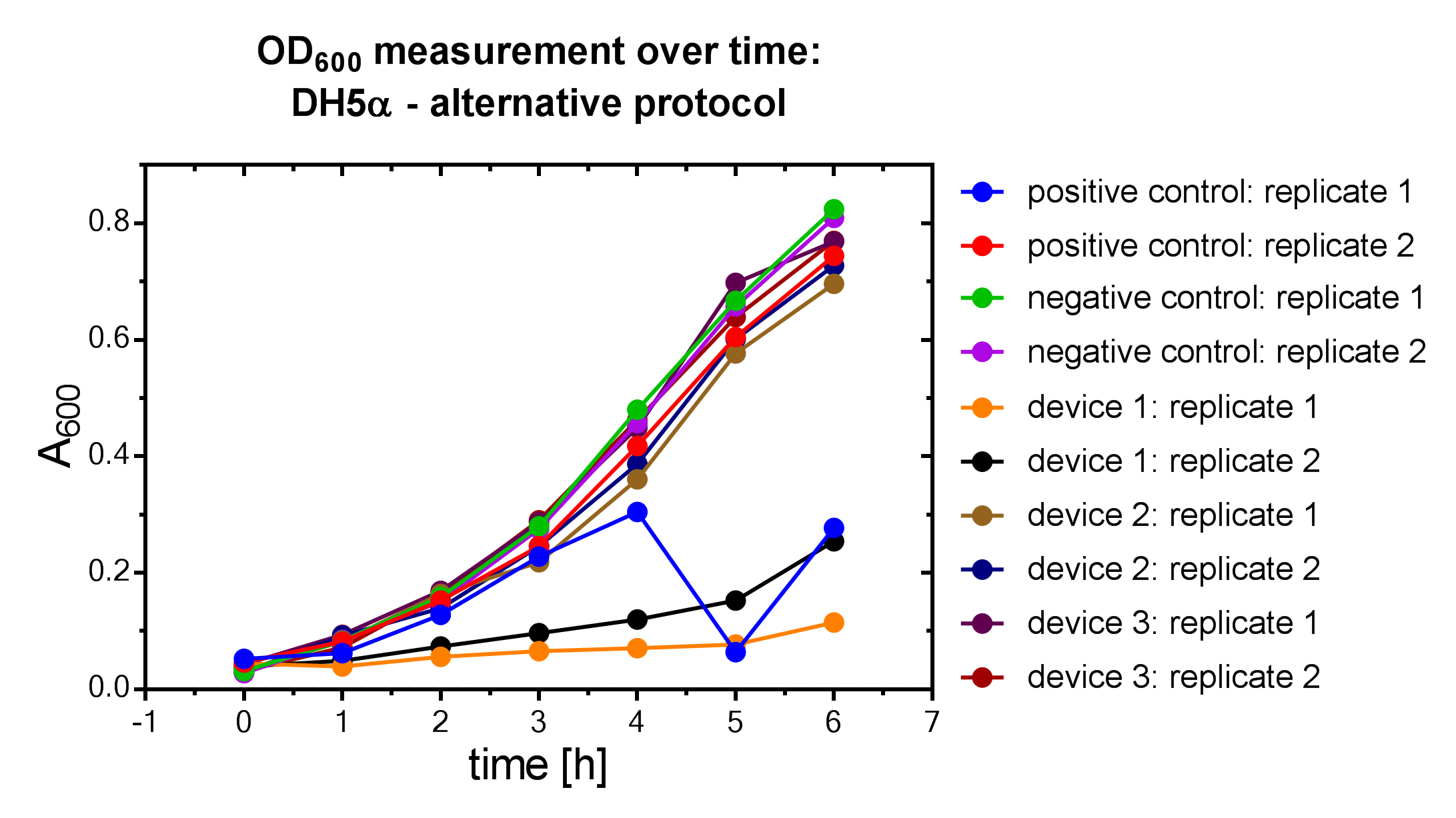
| + | File:Muc16_Interlab_od_alternative.png| <i><b>Figure 6:</b></i> Alternative OD<sub>600</sub> measurement of DH5α <i>E. coli </i> strain. | ||
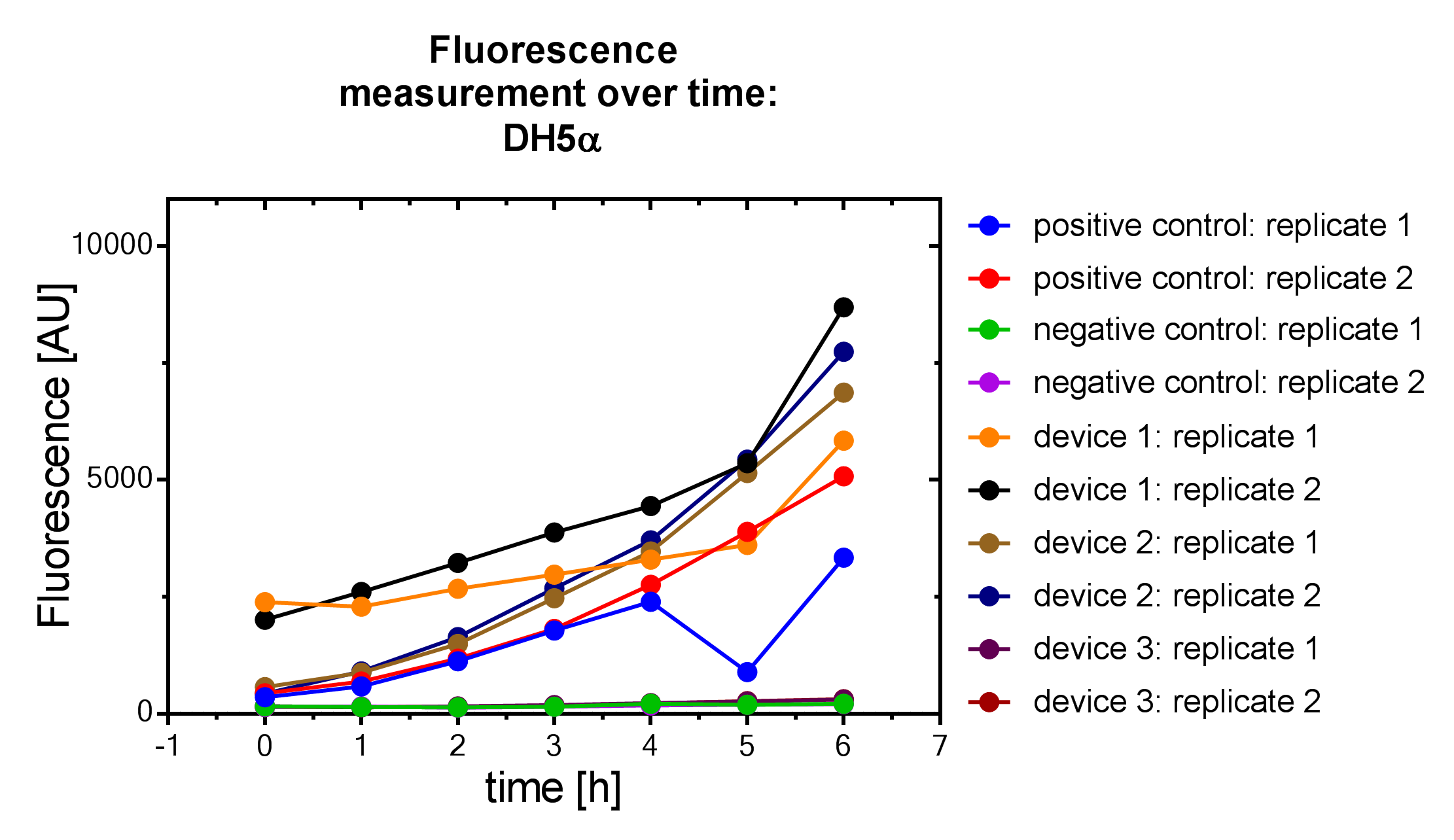
| + | File:Muc16_Interlab_fl_alternative.png| <i><b>Figure 7:</b></i> Alternative fluorescence measurement of DH5α <i>E. coli </i> strain. | ||
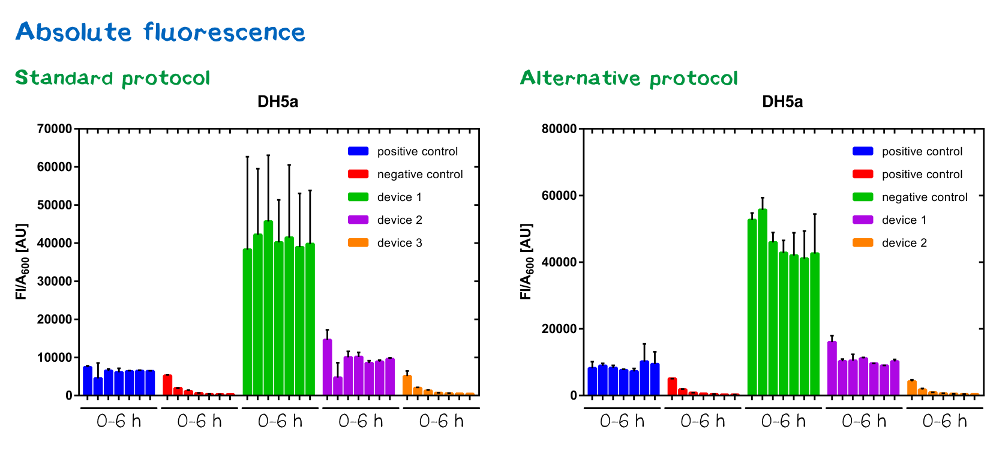
| + | File:Muc16_Interlab_abs_standard_alternative.png| <i><b>Figure 8:</b></i> Absolute fluorescence of DH5α <i>E. coli for standard and multi-measurement protocol</i> strain. | ||
| + | </gallery> | ||
| + | <br> | ||
| + | <!-- | ||
| + | [[File:Muc16_Interlab_od_protocol.png|thumb|right|250px| <i><b>Figure 4:</b></i> OD<sub>600</sub> measurement of DH5α <i>E. coli </i> strain.]] | ||
| + | [[File:Muc16_Interlab_fl_protocol.png|thumb|right|250px| <i><b>Figure 5:</b></i> Fluorescence measurement of DH5α <i>E. coli </i> strain.]] | ||
| + | [[File:Muc16_Interlab_od_alternative.png|thumb|right|250px| <i><b>Figure 6:</b></i> Alternative OD<sub>600</sub> measurement of DH5α <i>E. coli </i> strain.]] | ||
| + | [[File:Muc16_Interlab_fl_alternative.png|thumb|center|500px| <i><b>Figure 7:</b></i> Alternative fluorescence measurement of DH5α <i>E. coli </i> strain.]] | ||
| − | + | <gallery> | |
| − | <gallery | + | File: Muc16_Interlab_10beta.jpg | ''OD<sub>600</sub> measurement: 10β'' |
| − | File:Muc16_Interlab_10beta.jpg| ''OD<sub>600</sub> measurement: 10β'' | + | |
File:Muc16_Interlab_jm83.jpg| ''OD<sub>600</sub> measurement: jm83 blue'' | File:Muc16_Interlab_jm83.jpg| ''OD<sub>600</sub> measurement: jm83 blue'' | ||
File:Muc16_Interlab_ks272.jpg| ''OD<sub>600</sub> measurement: ks272'' | File:Muc16_Interlab_ks272.jpg| ''OD<sub>600</sub> measurement: ks272'' | ||
| Line 58: | Line 58: | ||
</gallery> | </gallery> | ||
--> | --> | ||
| + | </div> | ||
| + | |||
| + | <!--- | ||
| + | ==Random stuff from the lab== | ||
| + | <ul class="gallery"> | ||
| + | <li>[[Media:Muc16_Interlab_10beta.jpg]][[Image:Muc16_Interlab_10beta.jpg|200px|Tests|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_2.jpeg]][[Image:Muc16_Gallery_2.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_3.jpeg]][[Image:Muc16_Gallery_3.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_4.jpeg]][[Image:Muc16_Gallery_4.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_6.jpeg]][[Image:Muc16_Gallery_6.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_7.jpeg]][[Image:Muc16_Gallery_7.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_8.jpeg]][[Image:Muc16_Gallery_8.jpeg|200px|link=]]</li> | ||
| + | <li>[[Media:Muc16_Gallery_9.jpeg]][[Image:Muc16_Gallery_9.jpeg|200px|link=]]</li> | ||
| + | </ul> | ||
| + | ---> | ||
| − | == | + | ==Discussion== |
| − | Due to the new protocol it is possible to measure multiple samples simultaneously by only one person. Also less space is needed to carry out the experiment due to the compact 96 Masterblock. Besides of this the instant | + | <div class="white-box"> |
| + | Due to the new protocol, it is possible to measure multiple samples simultaneously by only one person. Also, less space is needed to carry out the experiment due to the compact 96 Masterblock. Besides of this the instant measurement of the samples, taken every hour, is much more accurate than keeping them on ice for one single end measurement. As you can see on the data above there are still some problems left like unnoticed air bubbles in some wells which obviously leads to outliers in the dataset. Also slightly different volumina in the wells can lead to some fluctuation in the datasets. | ||
All in all it seems that the use of different strains had no effect on the results(data not shown), just as it was postulated before.<ref>Beal, J., Haddock-Angelli, T., Gershater, M., de Mora, K., Lizarazo, M., Hollenhorst, J., & Rettberg, R. (2016). Reproducibility of Fluorescent Expression from Engineered Biological Constructs in E. coli. PloS one, 11(3), e0150182.</ref>. | All in all it seems that the use of different strains had no effect on the results(data not shown), just as it was postulated before.<ref>Beal, J., Haddock-Angelli, T., Gershater, M., de Mora, K., Lizarazo, M., Hollenhorst, J., & Rettberg, R. (2016). Reproducibility of Fluorescent Expression from Engineered Biological Constructs in E. coli. PloS one, 11(3), e0150182.</ref>. | ||
Never the less we hope that the data created by use of this advanced multi-measurement protocol will help to furthermore proof this assumption. | Never the less we hope that the data created by use of this advanced multi-measurement protocol will help to furthermore proof this assumption. | ||
| + | </div> | ||
| + | ==References== | ||
| + | <div class="white-box"> | ||
| + | <references /> | ||
| + | </div> | ||
{{LMU-TUM_Munich_html_end}} | {{LMU-TUM_Munich_html_end}} | ||
Latest revision as of 03:33, 20 October 2016
InterLab
For the purpose of the third interlaboratory measurement study organized by iGEM about the reproducibility of fluorescence measurements, five different GFP expression plasmids were transformed and expressed in the growth rate of E. coli bacteria. The corresponding expression levels of the different constructs were determined by measuring OD600 and fluorescence by a plate reader over time.
Introduction
Science is only as good as the experiments underlying it - and experiments are only as good as the accuracy of their measurements. Once again, iGEM has called labs all around the world to their instruments, in order to determine if a seemingly same experiment really is the same experiment everywhere - or whether the differences in instrument architecture and accuracy may make reproducibility a problem. For the third iGEM interlab study, fluorescence measurements with five different GFP expression plasmids in E. coli were performed. The growth rate of the bacteria and the corresponding expression levels of the different GFP constructs were determined via a plate reader. We are glad to be able to do our part for this crucial issue, especially since the competition is such a great place to gather data from all around the world!
Materials & Methods
Materials
- [http://parts.igem.org/Help:InterLab_Measurement_Kit InterLab Measurement Kit]
- Plate reader: [http://www.biotek.com/products/microplate_detection/synergy2_multimode_microplate_reader.html?utm_source=bing&utm_medium=cpc&utm_campaign=Readers&utm_term=biotek%20synergy%202&utm_content=Microplate%20Readers%20-%20Synergy Biotec Sinergy 2]
- Microtest plate: Microtest Plate 96 Well,F; Sarstedt; order number: 82.1581
- Masterblock: MASTERBLOCK®, 96 WELL, 2 ML, PP, V-BOTTOM, NATURAL, SINGLE PACKED; greiner bio-one; item number: 780270
- E. coli strains: DH5α, W3110, KS272, XL-1 blue, JM83, OriB, 10β NEB turbo
Methods
Given protocol
- According to provided plate reader protocol
Multi-measurement protocol
All five plasmids were transformed in different E. coli strains. Single colonies were picked and grown at 220 rpm and 37 °C in 1 mL LB liquid culture per well in one Masterblock over night. The layout of the plate can be found in figure 1 and figure 3. On the next day OD600 were measured by pipetting 100 µL of each well into a microtest plate with the same layout using a 12 channel pipette. According to the results new diluted LB liquid cultures were mixed in a new Masterblock with the same layout and incubated at 37 °C for 6 hours at 220 rpm. At the beginning and every hour a sample was taken by transferring 100 µL of each well into a new microtest plate with the same layout, see figure 2.
Results
Samples were prepared according to our multi-measurement protocol and the standard protocol. The results for DH5α E. coli strain are plotted in figure 4-8.
Discussion
Due to the new protocol, it is possible to measure multiple samples simultaneously by only one person. Also, less space is needed to carry out the experiment due to the compact 96 Masterblock. Besides of this the instant measurement of the samples, taken every hour, is much more accurate than keeping them on ice for one single end measurement. As you can see on the data above there are still some problems left like unnoticed air bubbles in some wells which obviously leads to outliers in the dataset. Also slightly different volumina in the wells can lead to some fluctuation in the datasets.
All in all it seems that the use of different strains had no effect on the results(data not shown), just as it was postulated before.[1].
Never the less we hope that the data created by use of this advanced multi-measurement protocol will help to furthermore proof this assumption.
References
- ↑ Beal, J., Haddock-Angelli, T., Gershater, M., de Mora, K., Lizarazo, M., Hollenhorst, J., & Rettberg, R. (2016). Reproducibility of Fluorescent Expression from Engineered Biological Constructs in E. coli. PloS one, 11(3), e0150182.