m |
(Add biological capacitor description.) |
||
| Line 1: | Line 1: | ||
| − | |||
<html> | <html> | ||
| − | |||
<div class="column"> | <div class="column"> | ||
<h1>Parts</h1> | <h1>Parts</h1> | ||
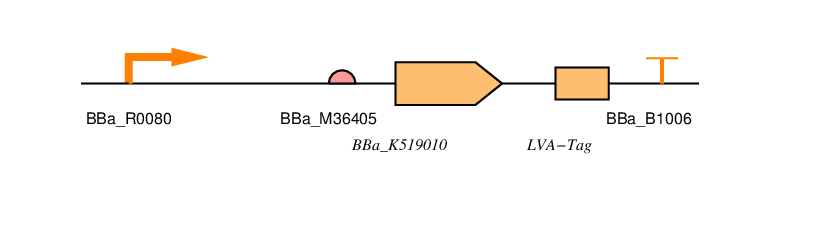
<p>You can also find all the parts we've designed in <a href="http://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2016&group=Newcastle">the parts registry</a>. We've used <a href="http://sbolstandard.org/visual/">SBOL visual</a> to specify our designs. <!-- TODO: Why? --></p> | <p>You can also find all the parts we've designed in <a href="http://parts.igem.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2016&group=Newcastle">the parts registry</a>. We've used <a href="http://sbolstandard.org/visual/">SBOL visual</a> to specify our designs. <!-- TODO: Why? --></p> | ||
<h2>Part Information</h2> | <h2>Part Information</h2> | ||
| − | <h3> | + | <div id="bio-capacitor"> |
| + | <h3>Biological 'Capacitor'</h3> | ||
| + | <p>We plan to engineer Escherichia coli to mimic one of the properties of a capacitor, the ability to accumulate and hold charge for some time before discharging. This is shown in the idealised graph below.</p> | ||
| + | |||
| + | <p><img title="" alt="Idealised capacitor charge-discharge cycle" src="https://static.igem.org/mediawiki/2016/a/ac/T--Newcastle--Capacitor-C-DC.png"></p> | ||
| + | |||
| + | <p>An electrical capacitor accumulates charge whilst a voltage is applied and then discharges when the voltage stops being applied. We make an analogy between the voltage signal and protein concentration. Whereas an electrical capacitor accumulates charge, a biological ‘capacitor’ would accumulate proteins. Like an electrical capacitor which has a maximum charge it can accumulate, there is a maximum protein concentration that can accumulate in the cell determined by its production and degradation rate. Once proteins stop being accumulated in the cell it ‘discharges’ by having these drive the production of an output signal.</p> | ||
| + | |||
| + | <p>In this construct we use L-arabinose to mimic a voltage signal. This is entirely for experimental purposes, there is no reason that this device cannot be modified to respond to an electrical signal, for instance through the heat shock response explored elsewhere in our work. Although in this case we show that protein’s can be accumulated there is no reason why actual charge, in the form of a potential difference could not be generated across the cell membrane. There are already examples of <a href="https://en.wikipedia.org/wiki/Membrane_potential">membrane potentials</a> in biology, the most obvious being found in neurons. This is something that has been explored by iGEM teams in the past e.g., <a href="https://2008.igem.org/IGEM:Cambridge/2008/Notebook/Voltage/K%2B_Concentrations">Cambridge (2008)</a>. Importantly we show through modelling that the charge-discharge cycle can be mimicked in biological cells through the use of repressor/inducer competition. This could be merged with work on membrane potentials in the future. </p> | ||
| + | |||
| + | <p>Because we use a constitutively on promoter, the TetR repressible promoter (<a href="http://parts.igem.org/Part:BBa_R0040">BBa_R0040</a>) the default state of the system is ‘charging’. In this state lambda repressor (<a href="http://parts.igem.org/Part:BBa_C1051">BBa_C1051</a>) accumulates in the cell together with 434 repressor (<a href="http://parts.igem.org/Part:BBa_C0052">BBa_C0052</a>). The amount of 434 repressor grows faster than that of lambda repressor because there are two coding sequences for the protein in the circiut. This is to ensure that it outcompetes (on average) the lambda repressor so that there is a low output signal whilst in the charging state. This occurs because 434 repressor represses the output promoter whilst lambda repressor induces it. In out device the output signal is sfGFP (<a href="http://parts.igem.org/Part:BBa_I746916">BBa_I746916</a>).</p> | ||
| + | |||
| + | <p>We can switch the state of the system to the discharging state by causing the expression of TetR. To facilitate this we have used an L-arabinose promoter coupled with the TetR coding sequence to give us a chemical ‘off switch’. Once the TetR is produced the system enters the discharging state, no further protein synthesis in our construct is induced and so the amount of 434 repressor and lambda repressor start to decay. The 434 repressor is tagged with a very fast <a href="http://parts.igem.org/Protein_domains/Degradation">ssRA degradation tag</a>, the LVA degradation tag so that it will be broken down faster than lambda repressor.</p> | ||
| + | |||
| + | <p>As this happens there will reach a point where the 434 repressor stops out-competing the lambda repressor and the output will start to be produced as it is induced by the lambda repressor. Whilst this is happening the level of 434 and lambda repressor will continue to fall until the output stops being produced and the system has completely ‘discharged’ and is in a resting state. At this point the removal of L-arabinose and the addition of tetracycline or an analogue thereof (which binds to TetR and prevents it from repressing the promoter) would switch the system back into the charging state and the process can begin again.</p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<img src="https://static.igem.org/mediawiki/2016/e/e3/T--Newcastle--LightBulb.png" /> | <img src="https://static.igem.org/mediawiki/2016/e/e3/T--Newcastle--LightBulb.png" /> | ||
<h3>Variable Resistor</h3> | <h3>Variable Resistor</h3> | ||
Revision as of 10:38, 12 August 2016
Parts
You can also find all the parts we've designed in the parts registry. We've used SBOL visual to specify our designs.
Part Information
Biological 'Capacitor'
We plan to engineer Escherichia coli to mimic one of the properties of a capacitor, the ability to accumulate and hold charge for some time before discharging. This is shown in the idealised graph below.

An electrical capacitor accumulates charge whilst a voltage is applied and then discharges when the voltage stops being applied. We make an analogy between the voltage signal and protein concentration. Whereas an electrical capacitor accumulates charge, a biological ‘capacitor’ would accumulate proteins. Like an electrical capacitor which has a maximum charge it can accumulate, there is a maximum protein concentration that can accumulate in the cell determined by its production and degradation rate. Once proteins stop being accumulated in the cell it ‘discharges’ by having these drive the production of an output signal.
In this construct we use L-arabinose to mimic a voltage signal. This is entirely for experimental purposes, there is no reason that this device cannot be modified to respond to an electrical signal, for instance through the heat shock response explored elsewhere in our work. Although in this case we show that protein’s can be accumulated there is no reason why actual charge, in the form of a potential difference could not be generated across the cell membrane. There are already examples of membrane potentials in biology, the most obvious being found in neurons. This is something that has been explored by iGEM teams in the past e.g., Cambridge (2008). Importantly we show through modelling that the charge-discharge cycle can be mimicked in biological cells through the use of repressor/inducer competition. This could be merged with work on membrane potentials in the future.
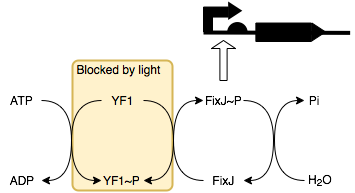
Because we use a constitutively on promoter, the TetR repressible promoter (BBa_R0040) the default state of the system is ‘charging’. In this state lambda repressor (BBa_C1051) accumulates in the cell together with 434 repressor (BBa_C0052). The amount of 434 repressor grows faster than that of lambda repressor because there are two coding sequences for the protein in the circiut. This is to ensure that it outcompetes (on average) the lambda repressor so that there is a low output signal whilst in the charging state. This occurs because 434 repressor represses the output promoter whilst lambda repressor induces it. In out device the output signal is sfGFP (BBa_I746916).
We can switch the state of the system to the discharging state by causing the expression of TetR. To facilitate this we have used an L-arabinose promoter coupled with the TetR coding sequence to give us a chemical ‘off switch’. Once the TetR is produced the system enters the discharging state, no further protein synthesis in our construct is induced and so the amount of 434 repressor and lambda repressor start to decay. The 434 repressor is tagged with a very fast ssRA degradation tag, the LVA degradation tag so that it will be broken down faster than lambda repressor.
As this happens there will reach a point where the 434 repressor stops out-competing the lambda repressor and the output will start to be produced as it is induced by the lambda repressor. Whilst this is happening the level of 434 and lambda repressor will continue to fall until the output stops being produced and the system has completely ‘discharged’ and is in a resting state. At this point the removal of L-arabinose and the addition of tetracycline or an analogue thereof (which binds to TetR and prevents it from repressing the promoter) would switch the system back into the charging state and the process can begin again.

Variable Resistor
Light Dependent Resistor
A

B
Time Delay
Each team will make new parts during iGEM and will submit them to the Registry of Standard Biological Parts. The iGEM software provides an easy way to present the parts your team has created. The <groupparts> tag (see below) will generate a table with all of the parts that your team adds to your team sandbox.
Remember that the goal of proper part documentation is to describe and define a part, so that it can be used without needing to refer to the primary literature. Registry users in future years should be able to read your documentation and be able to use the part successfully. Also, you should provide proper references to acknowledge previous authors and to provide for users who wish to know more.
Note
Note that parts must be documented on the Registry. This page serves to showcase the parts you have made. Future teams and other users and are much more likely to find parts by looking in the Registry than by looking at your team wiki.
Adding parts to the registry
You can add parts to the Registry at our Add a Part to the Registry link.
We encourage teams to start completing documentation for their parts on the Registry as soon as you have it available. The sooner you put up your parts, the better you will remember all the details about your parts. Remember, you don't need to send us the DNA sample before you create an entry for a part on the Registry. (However, you do need to send us the DNA sample before the Jamboree. If you don't send us a DNA sample of a part, that part will not be eligible for awards and medal criteria.)
What information do I need to start putting my parts on the Registry?
The information needed to initially create a part on the Registry is:
- Part Name
- Part type
- Creator
- Sequence
- Short Description (60 characters on what the DNA does)
- Long Description (Longer description of what the DNA does)
- Design considerations
We encourage you to put up much more information as you gather it over the summer. If you have images, plots, characterization data and other information, please also put it up on the part page.
Inspiration
We have a created a collection of well documented parts that can help you get started.
You can also take a look at how other teams have documented their parts in their wiki:

