(Prototype team page) |
|||
| Line 1: | Line 1: | ||
| − | |||
<html> | <html> | ||
| + | <head> | ||
| + | <link rel="icon" type="image/png" href="https://static.igem.org/mediawiki/2016/f/f5/T-Peshawar--favicon-v1-compressor.png"> | ||
| + | <title>Modelling - iGEM Peshawar 2016</title> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2016.igem.org/Template:Peshawar/CSS?action=raw&ctype=text/css" /> | ||
| + | <script type="text/x-mathjax-config"> | ||
| + | MathJax.Hub.Config({ | ||
| + | tex2jax: { | ||
| + | inlineMath: [ ['$','$'], ["\\(","\\)"] ], | ||
| + | processEscapes: true | ||
| + | } | ||
| + | }); | ||
| + | </script> | ||
| + | <script type="text/javascript" src="http://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"></script> | ||
| − | + | </head> | |
| − | + | ||
| − | + | ||
| + | <body> | ||
| + | <div class="outermost-box"> | ||
| + | <div id="menu-button" class="floating-button-wrapper"> | ||
| + | <div id="menu-background" class="js-toggle-left-slidebar"> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!--End of floating menu button wrapper --> <div off-canvas="slidebar-1 left reveal"> | ||
| + | <nav> | ||
| + | <ul class="navigation-list"> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar">Home</a></li> | ||
| + | <hr> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Description">Project</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Parts">Parts</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Model">Modeling</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Safety">Safety</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Notebook">Notebook</a></li> | ||
| + | <hr> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Team">Team</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Attributions">Attributions</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Collaborations">Collaborations</a></li> | ||
| + | <hr> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Human_Practices">Human Practices</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/HP/Silver">Silver</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Engagement">Public Engagement</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Awards">Awards</a></li> | ||
| + | <hr> | ||
| + | <li><a href="https://2016.igem.org/Team:Peshawar/Wiki">How to design an iGEM Wiki</a></li> | ||
| − | < | + | </ul> |
| + | </nav> | ||
| + | <article id="social-main-footer"> | ||
| + | <!--FACEBOOK--> | ||
| + | <a href="https://www.facebook.com/iGEMPeshawar2016/" target="_blank"> | ||
| + | <svg class="social-main-footer-svg facebook" width="61" height="60" viewBox="0 0 61 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--facebook-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--facebook-round-v2" fill="#808184"> | ||
| + | <path d="M30.594 1.465c-15.735 0-28.536 12.8-28.536 28.537 0 15.734 12.8 28.536 28.536 28.536S59.13 45.736 59.13 30.002c0-15.736-12.8-28.537-28.536-28.537m0 58.536c-16.542 0-30-13.455-30-29.997 0-16.543 13.458-30 30-30s30 13.457 30 30c0 16.542-13.458 30-30 30" id="facebook-circle" /> | ||
| + | <path d="M22.91 25.905h3.72V22.29c0-1.597.042-4.057 1.2-5.58 1.22-1.614 2.895-2.71 5.777-2.71 4.696 0 6.672.67 6.672.67l-.933 5.514s-1.55-.45-2.997-.45c-1.447 0-2.743.52-2.743 1.966v4.205h5.936l-.414 5.386h-5.524V50H26.63V31.29h-3.72v-5.385z" id="facebook" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--TWITTER--> | ||
| + | <a href="https://twitter.com/igem_peshawar" target="_blank"> | ||
| + | <svg class="social-main-footer-svg twitter" width="60" height="60" viewBox="0 0 60 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--twitter-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--twitter-round-v2" fill="#808184"> | ||
| + | <path d="M30 1.463C14.266 1.463 1.464 14.265 1.464 30S14.264 58.537 30 58.537 58.538 45.735 58.538 30 45.738 1.463 30 1.463M30 60C13.46 60 0 46.542 0 30S13.46 0 30 0c16.542 0 30 13.458 30 30S46.542 60 30 60" id="twitter-circle" /> | ||
| + | <path d="M49.642 21.783c-1.325.587-2.75.984-4.242 1.163 1.527-.914 2.698-2.36 3.247-4.085-1.428.848-3.006 1.46-4.692 1.79-1.343-1.433-3.265-2.33-5.39-2.33-4.077 0-7.384 3.308-7.384 7.385 0 .58.065 1.142.19 1.683-6.134-.31-11.576-3.247-15.222-7.718-.636 1.093-.998 2.362-.998 3.715 0 2.56 1.303 4.82 3.283 6.148-1.208-.038-2.35-.373-3.347-.923v.09c0 3.58 2.547 6.566 5.927 7.242-.62.174-1.27.26-1.946.26-.476 0-.94-.044-1.39-.13.94 2.933 3.666 5.07 6.9 5.126-2.53 1.982-5.715 3.163-9.173 3.163-.596 0-1.185-.032-1.762-.1 3.27 2.093 7.15 3.316 11.32 3.316 13.59 0 21.015-11.25 21.015-21.013 0-.32-.006-.64-.02-.955 1.444-1.043 2.7-2.344 3.685-3.824" id="twitter" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--INSTAGRAM--> | ||
| + | <a href="https://www.instagram.com/igempeshawar/" target="_blank"> | ||
| + | <svg class="social-main-footer-svg instagram" width="61" height="61" viewBox="0 0 61 61" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--instagram-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--instagram-round-v2" fill="#808184"> | ||
| + | <path d="M30.802 1.64c-15.736 0-28.537 12.802-28.537 28.537s12.8 28.537 28.537 28.537c15.735 0 28.536-12.802 28.536-28.537S46.538 1.64 30.802 1.64m0 58.537c-16.542 0-30-13.458-30-30s13.458-30 30-30 30 13.458 30 30-13.458 30-30 30" id="instagram-circle" /> | ||
| + | <path d="M45.3 42.212c0 1.902-1.548 3.446-3.45 3.446H19.763c-1.903 0-3.45-1.544-3.45-3.446v-14.73h5.375c-.464 1.144-.727 2.39-.727 3.695 0 5.426 4.417 9.837 9.845 9.837 5.428 0 9.843-4.41 9.843-9.837 0-1.306-.264-2.55-.725-3.694H45.3v14.73zm-19.63-14.73c1.15-1.593 3.022-2.64 5.137-2.64 2.115 0 3.985 1.047 5.136 2.64.75 1.042 1.203 2.314 1.203 3.695 0 3.492-2.845 6.33-6.34 6.33-3.494 0-6.336-2.838-6.336-6.33 0-1.38.45-2.653 1.2-3.694zm18.174-10.145l.795-.004v6.097l-6.083.022-.022-6.098 5.308-.017zM41.85 13.19H19.763c-3.835 0-6.957 3.118-6.957 6.952v22.07c0 3.834 3.122 6.952 6.957 6.952H41.85c3.834 0 6.957-3.118 6.957-6.952v-22.07c0-3.834-3.123-6.952-6.958-6.952z" id="instagram" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--BLOG--> | ||
| + | <a href="http://www.synbiokp.com/blog/" target="_blank"> | ||
| + | <svg class="social-main-footer-svg blog" width="60" height="60" viewBox="0 0 60 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--blog-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--blog-round-v2" fill="#808184"> | ||
| + | <path d="M30 1.463C14.265 1.463 1.463 14.263 1.463 30c0 15.735 12.802 28.536 28.537 28.536S58.537 45.736 58.537 30 45.735 1.463 30 1.463M0 30C0 13.458 13.458 0 30 0s30 13.458 30 30-13.458 30-30 30S0 46.542 0 30z" id="blog-circle" /> | ||
| + | <path d="M34.95 19.916c7.654 0 14 6.357 14 14.025 0 7.64-6.375 14.03-14 14.03h-7.942C19.393 47.968 13 41.57 13 33.94V15.22c0-1.78 1.47-3.255 3.25-3.255 1.768 0 3.247 1.482 3.247 3.256 0 6.24.01 12.48.01 18.72 0 4.11 3.4 7.52 7.5 7.52h7.943c4.084 0 7.504-3.424 7.504-7.514 0-4.108-3.403-7.516-7.504-7.516h-8.568c-1.777 0-3.248-1.475-3.248-3.255 0-1.773 1.477-3.254 3.248-3.254h8.568z" id="blog" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | </article> | ||
</div> | </div> | ||
| + | <!--End of off-canvas menu--> <div canvas="container"> | ||
| + | <section class="inner-container"> | ||
| + | <div id="header-oc" class="header-outer-container"> | ||
| + | <div class="sticky-header"> | ||
| + | <div class="logo-wrapper"> | ||
| + | <a href="https://2016.igem.org/Team:Peshawar"> | ||
| + | <img id="id-logo" class="logo" src="https://static.igem.org/mediawiki/2016/7/7b/T--Peshawar--iGEM-Peshawar-Logo---New-Pakol-v2-2.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!--End of full width header containing responsive logo --> <script type="text/javascript"> | ||
| + | |||
| + | var button = document.getElementById('menu-button'); | ||
| + | button.style.position = "fixed"; | ||
| + | button.style.zIndex = "-1"; | ||
| + | var buttonImage = document.getElementById("menu-background"); | ||
| + | |||
| + | //Wait for the Logo to load and fill he div | ||
| + | document.getElementById('id-logo').onload = function() { | ||
| + | var header = document.getElementById('header-oc'); | ||
| + | headerHeight = header.scrollHeight; | ||
| + | screenWidth = header.scrollWidth; | ||
| + | console.log('headerHeight',headerHeight, 'headerwidth', screenWidth); | ||
| + | |||
| + | //Scale the button according to header height | ||
| + | var buttonheight = headerHeight / 3.5; | ||
| + | buttonImage.style.backgroundSize = buttonheight + 'px'; | ||
| + | buttonImage.style.height = buttonheight +'px'; | ||
| + | buttonImage.style.width = buttonheight*1.25 +'px'; | ||
| + | button.style.zIndex = "2"; | ||
| + | |||
| + | // Calculate button coordinates | ||
| + | menuButtonHeight = 80; | ||
| + | var buttonTopPosition = headerHeight / 2 - buttonheight / 2; | ||
| + | var buttonLeftPosition = screenWidth / 36 ; | ||
| + | |||
| + | // Poistion the button at optimal location | ||
| + | button.style.left = buttonLeftPosition + 'px'; | ||
| + | button.style.top = buttonTopPosition + 'px'; | ||
| + | }; | ||
| + | |||
| + | // Hack for menu button not appear when the back button is pressed in history | ||
| + | window.onpageshow = function(event) { | ||
| + | var button = document.getElementById('menu-button'); | ||
| + | button.style.position = "fixed"; | ||
| + | button.style.zIndex = "-1"; | ||
| + | var buttonImage = document.getElementById("menu-background"); | ||
| + | |||
| + | var header = document.getElementById('header-oc'); | ||
| + | headerHeight = header.scrollHeight; | ||
| + | screenWidth = header.scrollWidth; | ||
| + | console.log('headerHeight',headerHeight, 'headerwidth', screenWidth); | ||
| + | |||
| + | //Scale the button according to header height | ||
| + | var buttonheight = headerHeight / 3.5; | ||
| + | buttonImage.style.backgroundSize = buttonheight + 'px'; | ||
| + | buttonImage.style.height = buttonheight +'px'; | ||
| + | buttonImage.style.width = buttonheight*1.25 +'px'; | ||
| + | button.style.zIndex = "2"; | ||
| + | |||
| + | // Calculate button coordinates | ||
| + | menuButtonHeight = 80; | ||
| + | var buttonTopPosition = headerHeight / 2 - buttonheight / 2; | ||
| + | var buttonLeftPosition = screenWidth / 36 ; | ||
| + | |||
| + | // Poistion the button at optimal location | ||
| + | button.style.left = buttonLeftPosition + 'px'; | ||
| + | button.style.top = buttonTopPosition + 'px'; | ||
| + | }; | ||
| + | |||
| + | </script> | ||
| + | <div class="full-content-wrapper-container"> | ||
| + | <div class="full-width-content-wrapper"> | ||
| + | |||
| + | </div> | ||
| + | <!--End of full-width-content-wrapper--> | ||
| + | </div> | ||
| + | |||
| + | <div class="centered-content-wrapper-container team-container heading"> | ||
| + | <div class="centered-content-wrapper team-wrapper"> | ||
| + | <h2>Modelling (Under Construction)</h2> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper--> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper-container--> | ||
| + | |||
| + | <div class="centered-content-wrapper-container"> | ||
| + | <div class="centered-content-wrapper"> | ||
| + | <div> | ||
| + | <a href="https://plot.ly/~adnanniazi/8/" target="_blank" title="mCP Expression" style="display: block; text-align: center;"><img src="https://plot.ly/~adnanniazi/8.png" alt="<b></b>" style="max-width: 100%;width: 840px;" width="840" onerror="this.onerror=null;this.src='https://plot.ly/404.png';" /></a> | ||
| + | <script data-plotly="adnanniazi:8" src="https://plot.ly/embed.js" async></script> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper-container--> | ||
| + | |||
| + | |||
| + | <div class="centered-content-wrapper-container"> | ||
| + | <div class="centered-content-wrapper"> | ||
| + | <div> | ||
| + | <a href="https://plot.ly/~adnanniazi/14/" target="_blank" title="Chromoprotein" style="display: block; text-align: center;"><img src="https://plot.ly/~adnanniazi/14.png" alt="<b></b>" style="max-width: 100%;width: 840px;" width="840" onerror="this.onerror=null;this.src='https://plot.ly/404.png';" /></a> | ||
| + | <script data-plotly="adnanniazi:14" src="https://plot.ly/embed.js" async></script> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper-container--> | ||
| + | |||
| + | |||
| + | <!--End of full-width-content-wrapper-container--> | ||
| + | <div class="centered-content-wrapper-container"> | ||
| + | <div class="centered-content-wrapper"> | ||
| + | $$\mathbf{[CO]+[CooA] \longleftrightarrow [(CooA)2CO]}$$ | ||
| + | $$\mathbf{[(CooA)2CO] \longleftrightarrow [(CooA)2CO.Pr]}$$ | ||
| + | |||
| + | |||
| + | $k_{on1} = \textrm{formation rate constant}$ | ||
| + | <br> | ||
| + | $\gamma_1 = \textrm{degradation rate constant}$ | ||
| + | <br> | ||
| + | $P_{exp} = \textrm{probability of promoter to be activated}$ | ||
| + | |||
| + | $$P_{exp} = \frac{\textrm{number of situations in which the promoter will be activated}}{\textrm{total number of situations}} = \frac{w}{z}$$ | ||
| + | |||
| + | $$ w=\textrm{[(CooA)2CO.Pr]}=\frac{\textrm{[(CooA)2CO]}}{K_p}$$ | ||
| + | |||
| + | $$\frac{d(mRNA)}{dt}=k_{on1}.P_{exp}-\gamma_1.[mRNA]$$ | ||
| + | $$\textrm{Change in the concentration of the chromprotein} = \textrm{formation rate} - \textrm{degration rate}$$ | ||
| + | |||
| + | $$ z=w+1=\frac{\textrm{[(CooA)2CO]}}{K_p}+1$$ | ||
| + | $$\textrm{Total number of situations} = \textrm{Number of situations in which the promoter will be activated} - \textrm{Number of situations in which the promoter will not be acitvated}$$ | ||
| + | |||
| + | $$P_{exp} = \frac{w}{z} = \frac{\frac{\textrm{[(CooA)2CO]}}{K_p}}{\frac{\textrm{[(CooA)2CO]}}{K_p}+1}$$ | ||
| + | |||
| + | $$P_{exp} = \frac{\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} {1+\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} $$ | ||
| + | |||
| + | $$[mRNA]=\frac{k_{on1}.P_{exp}}{\gamma_1}(1-e^{-\gamma_1t})$$ | ||
| + | |||
| + | $$\frac{d\textrm{[CP]}}{dt} = k_{on2}.\textrm{[mRNA]}-\gamma_2\textrm{[CP]}$$ | ||
| + | |||
| + | $$\textrm{[CP]} = k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1.\gamma_2}(1-e^{-\gamma_2t}) + k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1(\gamma_1-\gamma_2)}(e^{-\gamma_2t}-e^{-\gamma_1t})$$ | ||
| + | |||
| + | <!--NOx circuit--> | ||
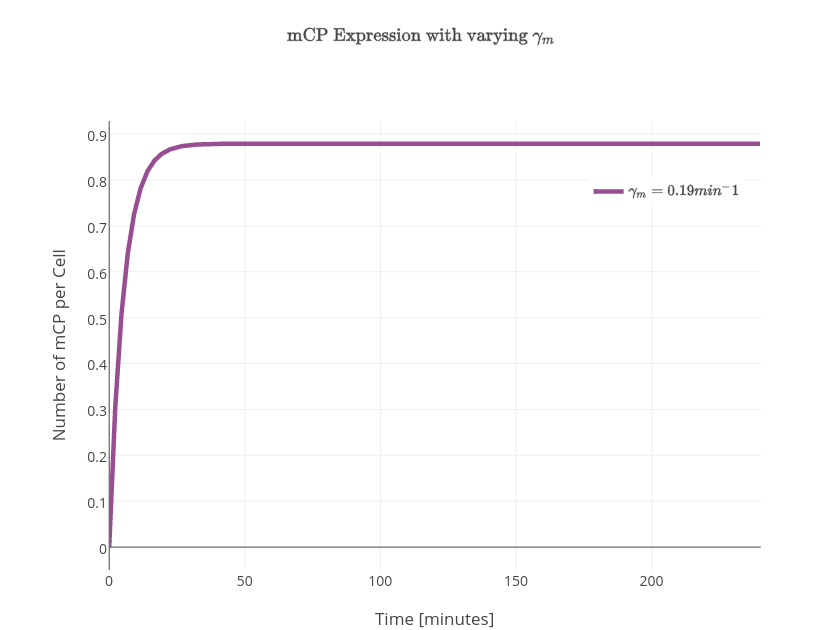
| + | $$\frac{d\textrm{[mCP]}}{dt} = k_{transcription}\textrm{[PYeaR]} - \gamma_m \textrm{[mCP]} $$ | ||
| + | |||
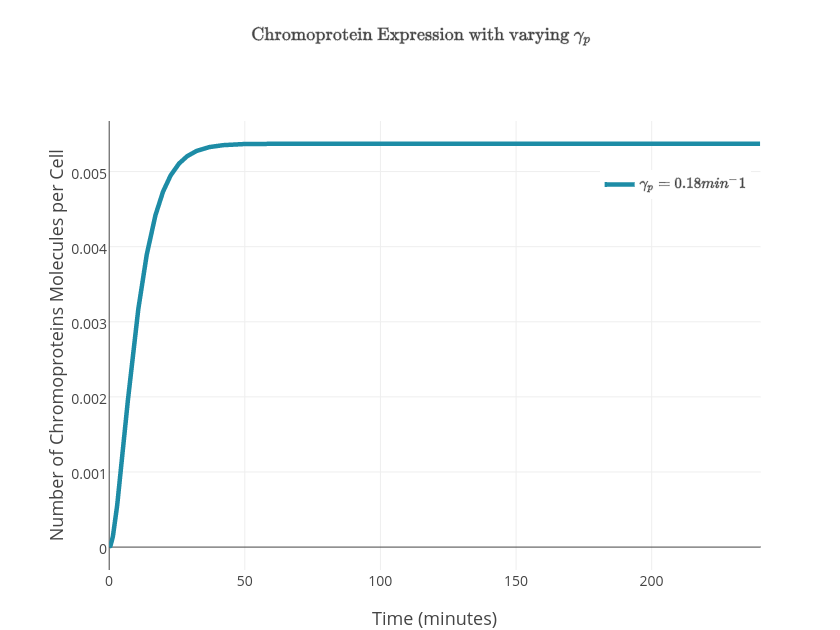
| + | $$\frac{d\textrm{[CP]}}{dt}=k_{translation}\textrm{[mCP]}-\gamma_p\textrm{[CP]}$$ | ||
| + | $$\frac{[\textrm{NO}^*]}{[\textrm{NO}_T]} = \frac{1}{1+\left(\frac{NsrR}{K_{NO}} \right)^n}$$ | ||
| + | $$\textrm{[PYeaR]}activity = \frac{\beta}{1+ \left( \frac{\textrm{NO}^*}{K_{d(\textrm{NsrR})}} \right)}$$ | ||
| + | $$\frac{d[\textrm{mCP}]}{dt} = \frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} } - \gamma_m [\textrm{mCP}]$$ | ||
| + | $$K=\frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} }$$ | ||
| + | $$\frac{d[\textrm{mCP}]}{dt} = K - \gamma_m [\textrm{mCP}]$$ | ||
| − | + | $$[\textrm{mCP}]=\frac{K}{\gamma_m} - \frac{K}{\gamma_m}e^{-\gamma_mt}$$ | |
| − | + | ||
| − | + | ||
| + | $$[\textrm{CP}]=\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} e^{-\gamma_mt} - \left(\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} \right)e^{-\gamma_pt} $$ | ||
| − | < | + | <table class="table-minimal"> |
| − | < | + | <thead> |
| − | + | <tr> | |
| − | </ | + | <th>Parameter </th> |
| − | < | + | <th>Description</th> |
| − | < | + | <th>Value</th> |
| − | < | + | </tr> |
| − | </ | + | </thead> |
| + | <tbody> | ||
| + | <tr> | ||
| + | <td>$[\textrm{PYeaR}]$</td> | ||
| + | <td>Concentration of PYeaR</td> | ||
| + | <td>1 copy/cell</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$k_{transcription}$</td> | ||
| + | <td>Rate of chromoprotein mRNA synthesis</td> | ||
| + | <td>0.167$\textrm{min}^{-1}$</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$k_{translation}$</td> | ||
| + | <td>Rate of chromoprotein synthesis</td> | ||
| + | <td>$0.0011\textrm{min}^{-1}$</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$\gamma_m$</td> | ||
| + | <td>mRNA degradation rate</td> | ||
| + | <td>$0.19\textrm{min}^{-1}$</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$\gamma_p$</td> | ||
| + | <td>Chromoprotein degradation rate</td> | ||
| + | <td>$0.18\textrm{min}^{-1}$</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$K_{\textrm{NO}}$</td> | ||
| + | <td>Dissociation constant of NO</td> | ||
| + | <td>$0.12\textrm{min}^{-1}$</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>$K_{\textrm{d(NsrR)}}$</td> | ||
| + | <td>Dissociation constant of NsrR</td> | ||
| + | <td>0.035 mM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>n</td> | ||
| + | <td>Cooperativity of NO binding to NsrR</td> | ||
| + | <td>2</td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <table class="table-minimal"> | ||
| + | <thead> | ||
| + | <tr> | ||
| + | <th>Header Cell 1</th> | ||
| + | <th>Header Cell 2</th> | ||
| + | <th>Header Cell 3</th> | ||
| + | </tr> | ||
| + | </thead> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td>Row 1 Cell 1</td> | ||
| + | <td>Row 1 Cell 2</td> | ||
| + | <td><button>Confirm</button><button>Reject</button></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Row 2 Cell 1</td> | ||
| + | <td>Row 2 Cell 2</td> | ||
| + | <td><button>Confirm</button><button>Reject</button></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Row 3 Cell 1</td> | ||
| + | <td>Row 3 Cell 2</td> | ||
| + | <td><button>Confirm</button><button>Reject</button></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Row 4 Cell 1</td> | ||
| + | <td>Row 4 Cell 2</td> | ||
| + | <td><button>Confirm</button><button>Reject</button></td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper--> | ||
| + | </div> | ||
| + | <!--End of centered-content-wrapper-container--> | ||
| + | |||
| + | <div class="all-in-one-footer-container"> | ||
| + | <div class="all-in-one-footer-address"> | ||
| + | <h3>Reach Us</h3> | ||
| + | <h4><img id="location" src="https://static.igem.org/mediawiki/2016/7/7c/T--Peshawar--location-compressor.png"> Institute of Integrative Biosciences, | ||
| + | <br>CECOS University of IT & Emerging Sciences, | ||
| + | <br>Phase 6, F-5, Hayatabad, Peshawar, Pakistan | ||
| + | </h4> | ||
| + | <h4 id="h4-email"><img id="email" src="https://static.igem.org/mediawiki/2016/0/04/T--Peshawar--email.png"> | ||
| + | <a href="mailto:iib@cecos.edu.pk">iib@cecos.edu.pk</a> | ||
| + | </h4> | ||
| + | <h4 id="h4-phone"><img id="phone" src="https://static.igem.org/mediawiki/2016/c/c6/T--Peshawar--phone-compressor.png"> +92 91 5860291</h4> | ||
| + | </div> | ||
| + | <div class="all-in-one-sponsors"> | ||
| + | <h3>Our Partners</h3> | ||
| + | <br> | ||
| + | <div class="sponsors-icons govt-kpk"> | ||
| + | <a href="http://kp.gov.pk/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/a/a1/T--Peshawar--govt-of-kpk-logo-small-gs-compressed.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="sponsors-icons dost"> | ||
| + | <a href="http://www.dost.gov.pk/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/e/e2/T--peshawar--dost-logo-small-gs-compresed-compressor.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="sponsors-icons cecos"> | ||
| + | <a href="http://cecos.edu.pk/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/0/03/T--Peshawar--cecos-logo-small-gs-compressed-compressor.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | <div class="sponsors-icons iib"> | ||
| + | <a href="http://iib.cecos.edu.pk/" target="_blank"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/6/60/T--Peshawar--iib-logo-small-gs-compressed-compressor.png"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="all-in-one-footer-social"> | ||
| + | <h3>Connect with us</h3> | ||
| + | <br> | ||
| + | <article id="social-page-footer"> | ||
| + | <!--FACEBOOK--> | ||
| + | <a href="https://www.facebook.com/iGEMPeshawar2016/" target="_blank"> | ||
| + | <svg class="social-page-footer-svg facebook" width="61" height="60" viewBox="0 0 61 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--facebook-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--facebook-round-v2" fill="#808184"> | ||
| + | <path d="M30.594 1.465c-15.735 0-28.536 12.8-28.536 28.537 0 15.734 12.8 28.536 28.536 28.536S59.13 45.736 59.13 30.002c0-15.736-12.8-28.537-28.536-28.537m0 58.536c-16.542 0-30-13.455-30-29.997 0-16.543 13.458-30 30-30s30 13.457 30 30c0 16.542-13.458 30-30 30" id="facebook-circle" /> | ||
| + | <path d="M22.91 25.905h3.72V22.29c0-1.597.042-4.057 1.2-5.58 1.22-1.614 2.895-2.71 5.777-2.71 4.696 0 6.672.67 6.672.67l-.933 5.514s-1.55-.45-2.997-.45c-1.447 0-2.743.52-2.743 1.966v4.205h5.936l-.414 5.386h-5.524V50H26.63V31.29h-3.72v-5.385z" id="facebook" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--TWITTER--> | ||
| + | <a href="https://twitter.com/igem_peshawar" target="_blank"> | ||
| + | <svg class="social-page-footer-svg twitter" width="60" height="60" viewBox="0 0 60 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--twitter-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--twitter-round-v2" fill="#808184"> | ||
| + | <path d="M30 1.463C14.266 1.463 1.464 14.265 1.464 30S14.264 58.537 30 58.537 58.538 45.735 58.538 30 45.738 1.463 30 1.463M30 60C13.46 60 0 46.542 0 30S13.46 0 30 0c16.542 0 30 13.458 30 30S46.542 60 30 60" id="twitter-circle" /> | ||
| + | <path d="M49.642 21.783c-1.325.587-2.75.984-4.242 1.163 1.527-.914 2.698-2.36 3.247-4.085-1.428.848-3.006 1.46-4.692 1.79-1.343-1.433-3.265-2.33-5.39-2.33-4.077 0-7.384 3.308-7.384 7.385 0 .58.065 1.142.19 1.683-6.134-.31-11.576-3.247-15.222-7.718-.636 1.093-.998 2.362-.998 3.715 0 2.56 1.303 4.82 3.283 6.148-1.208-.038-2.35-.373-3.347-.923v.09c0 3.58 2.547 6.566 5.927 7.242-.62.174-1.27.26-1.946.26-.476 0-.94-.044-1.39-.13.94 2.933 3.666 5.07 6.9 5.126-2.53 1.982-5.715 3.163-9.173 3.163-.596 0-1.185-.032-1.762-.1 3.27 2.093 7.15 3.316 11.32 3.316 13.59 0 21.015-11.25 21.015-21.013 0-.32-.006-.64-.02-.955 1.444-1.043 2.7-2.344 3.685-3.824" id="twitter" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--INSTAGRAM--> | ||
| + | <a href="https://www.instagram.com/igempeshawar/" target="_blank"> | ||
| + | <svg class="social-page-footer-svg instagram" width="61" height="61" viewBox="0 0 61 61" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--instagram-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--instagram-round-v2" fill="#808184"> | ||
| + | <path d="M30.802 1.64c-15.736 0-28.537 12.802-28.537 28.537s12.8 28.537 28.537 28.537c15.735 0 28.536-12.802 28.536-28.537S46.538 1.64 30.802 1.64m0 58.537c-16.542 0-30-13.458-30-30s13.458-30 30-30 30 13.458 30 30-13.458 30-30 30" id="instagram-circle" /> | ||
| + | <path d="M45.3 42.212c0 1.902-1.548 3.446-3.45 3.446H19.763c-1.903 0-3.45-1.544-3.45-3.446v-14.73h5.375c-.464 1.144-.727 2.39-.727 3.695 0 5.426 4.417 9.837 9.845 9.837 5.428 0 9.843-4.41 9.843-9.837 0-1.306-.264-2.55-.725-3.694H45.3v14.73zm-19.63-14.73c1.15-1.593 3.022-2.64 5.137-2.64 2.115 0 3.985 1.047 5.136 2.64.75 1.042 1.203 2.314 1.203 3.695 0 3.492-2.845 6.33-6.34 6.33-3.494 0-6.336-2.838-6.336-6.33 0-1.38.45-2.653 1.2-3.694zm18.174-10.145l.795-.004v6.097l-6.083.022-.022-6.098 5.308-.017zM41.85 13.19H19.763c-3.835 0-6.957 3.118-6.957 6.952v22.07c0 3.834 3.122 6.952 6.957 6.952H41.85c3.834 0 6.957-3.118 6.957-6.952v-22.07c0-3.834-3.123-6.952-6.958-6.952z" id="instagram" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | <!--BLOG--> | ||
| + | <a href="http://www.synbiokp.com/blog/" target="_blank"> | ||
| + | <svg class="social-page-footer-svg blog" width="60" height="60" viewBox="0 0 60 60" xmlns="http://www.w3.org/2000/svg"> | ||
| + | <title> | ||
| + | T--Peshawar--blog-round-v2 | ||
| + | </title> | ||
| + | <g id="Page-1" fill="none" fill-rule="evenodd"> | ||
| + | <g id="T--Peshawar--blog-round-v2" fill="#808184"> | ||
| + | <path d="M30 1.463C14.265 1.463 1.463 14.263 1.463 30c0 15.735 12.802 28.536 28.537 28.536S58.537 45.736 58.537 30 45.735 1.463 30 1.463M0 30C0 13.458 13.458 0 30 0s30 13.458 30 30-13.458 30-30 30S0 46.542 0 30z" id="blog-circle" /> | ||
| + | <path d="M34.95 19.916c7.654 0 14 6.357 14 14.025 0 7.64-6.375 14.03-14 14.03h-7.942C19.393 47.968 13 41.57 13 33.94V15.22c0-1.78 1.47-3.255 3.25-3.255 1.768 0 3.247 1.482 3.247 3.256 0 6.24.01 12.48.01 18.72 0 4.11 3.4 7.52 7.5 7.52h7.943c4.084 0 7.504-3.424 7.504-7.514 0-4.108-3.403-7.516-7.504-7.516h-8.568c-1.777 0-3.248-1.475-3.248-3.255 0-1.773 1.477-3.254 3.248-3.254h8.568z" id="blog" /> | ||
| + | </g> | ||
| + | </g> | ||
| + | </svg> | ||
| + | </a> | ||
| + | </article> | ||
| + | <div class="wiki-design"> | ||
| + | <!-- <h4>Wiki designed by <a href="https://2016.igem.org/Team:Peshawar/Team">Adnan Niazi</a></h4> --> | ||
| + | <br> | ||
| + | <button><a href="https://2016.igem.org/Team:Peshawar/Wiki">Learn iGEM Wiki design</a></button> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| + | <!--End of all-in-one-footer-container--> | ||
| + | </section> | ||
| + | <!--End of inner-container--> | ||
| + | </div> | ||
| + | <!--End of on-canvas container for all the content--> | ||
| + | </div> | ||
| + | <!--End of outermost box--> | ||
| + | <script type="text/javascript" src="https://2016.igem.org/Template:Peshawar/Javascript?action=raw&ctype=text/javascript"></script> | ||
| + | </body> | ||
</html> | </html> | ||
Revision as of 06:54, 12 October 2016
Modelling (Under Construction)
$$\mathbf{[CO]+[CooA] \longleftrightarrow [(CooA)2CO]}$$
$$\mathbf{[(CooA)2CO] \longleftrightarrow [(CooA)2CO.Pr]}$$
$k_{on1} = \textrm{formation rate constant}$
$\gamma_1 = \textrm{degradation rate constant}$
$P_{exp} = \textrm{probability of promoter to be activated}$ $$P_{exp} = \frac{\textrm{number of situations in which the promoter will be activated}}{\textrm{total number of situations}} = \frac{w}{z}$$ $$ w=\textrm{[(CooA)2CO.Pr]}=\frac{\textrm{[(CooA)2CO]}}{K_p}$$ $$\frac{d(mRNA)}{dt}=k_{on1}.P_{exp}-\gamma_1.[mRNA]$$ $$\textrm{Change in the concentration of the chromprotein} = \textrm{formation rate} - \textrm{degration rate}$$ $$ z=w+1=\frac{\textrm{[(CooA)2CO]}}{K_p}+1$$ $$\textrm{Total number of situations} = \textrm{Number of situations in which the promoter will be activated} - \textrm{Number of situations in which the promoter will not be acitvated}$$ $$P_{exp} = \frac{w}{z} = \frac{\frac{\textrm{[(CooA)2CO]}}{K_p}}{\frac{\textrm{[(CooA)2CO]}}{K_p}+1}$$ $$P_{exp} = \frac{\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} {1+\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} $$ $$[mRNA]=\frac{k_{on1}.P_{exp}}{\gamma_1}(1-e^{-\gamma_1t})$$ $$\frac{d\textrm{[CP]}}{dt} = k_{on2}.\textrm{[mRNA]}-\gamma_2\textrm{[CP]}$$ $$\textrm{[CP]} = k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1.\gamma_2}(1-e^{-\gamma_2t}) + k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1(\gamma_1-\gamma_2)}(e^{-\gamma_2t}-e^{-\gamma_1t})$$ $$\frac{d\textrm{[mCP]}}{dt} = k_{transcription}\textrm{[PYeaR]} - \gamma_m \textrm{[mCP]} $$ $$\frac{d\textrm{[CP]}}{dt}=k_{translation}\textrm{[mCP]}-\gamma_p\textrm{[CP]}$$ $$\frac{[\textrm{NO}^*]}{[\textrm{NO}_T]} = \frac{1}{1+\left(\frac{NsrR}{K_{NO}} \right)^n}$$ $$\textrm{[PYeaR]}activity = \frac{\beta}{1+ \left( \frac{\textrm{NO}^*}{K_{d(\textrm{NsrR})}} \right)}$$ $$\frac{d[\textrm{mCP}]}{dt} = \frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} } - \gamma_m [\textrm{mCP}]$$ $$K=\frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} }$$ $$\frac{d[\textrm{mCP}]}{dt} = K - \gamma_m [\textrm{mCP}]$$ $$[\textrm{mCP}]=\frac{K}{\gamma_m} - \frac{K}{\gamma_m}e^{-\gamma_mt}$$ $$[\textrm{CP}]=\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} e^{-\gamma_mt} - \left(\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} \right)e^{-\gamma_pt} $$
$\gamma_1 = \textrm{degradation rate constant}$
$P_{exp} = \textrm{probability of promoter to be activated}$ $$P_{exp} = \frac{\textrm{number of situations in which the promoter will be activated}}{\textrm{total number of situations}} = \frac{w}{z}$$ $$ w=\textrm{[(CooA)2CO.Pr]}=\frac{\textrm{[(CooA)2CO]}}{K_p}$$ $$\frac{d(mRNA)}{dt}=k_{on1}.P_{exp}-\gamma_1.[mRNA]$$ $$\textrm{Change in the concentration of the chromprotein} = \textrm{formation rate} - \textrm{degration rate}$$ $$ z=w+1=\frac{\textrm{[(CooA)2CO]}}{K_p}+1$$ $$\textrm{Total number of situations} = \textrm{Number of situations in which the promoter will be activated} - \textrm{Number of situations in which the promoter will not be acitvated}$$ $$P_{exp} = \frac{w}{z} = \frac{\frac{\textrm{[(CooA)2CO]}}{K_p}}{\frac{\textrm{[(CooA)2CO]}}{K_p}+1}$$ $$P_{exp} = \frac{\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} {1+\frac{\textrm{[CooA]}.\frac{\frac{\textrm{CO}}{K_I}}{1+\frac{\textrm{CO}}{K_I}}}{K_p}} $$ $$[mRNA]=\frac{k_{on1}.P_{exp}}{\gamma_1}(1-e^{-\gamma_1t})$$ $$\frac{d\textrm{[CP]}}{dt} = k_{on2}.\textrm{[mRNA]}-\gamma_2\textrm{[CP]}$$ $$\textrm{[CP]} = k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1.\gamma_2}(1-e^{-\gamma_2t}) + k_{on2}.\frac{k_{on1}.P_{exp}}{\gamma_1(\gamma_1-\gamma_2)}(e^{-\gamma_2t}-e^{-\gamma_1t})$$ $$\frac{d\textrm{[mCP]}}{dt} = k_{transcription}\textrm{[PYeaR]} - \gamma_m \textrm{[mCP]} $$ $$\frac{d\textrm{[CP]}}{dt}=k_{translation}\textrm{[mCP]}-\gamma_p\textrm{[CP]}$$ $$\frac{[\textrm{NO}^*]}{[\textrm{NO}_T]} = \frac{1}{1+\left(\frac{NsrR}{K_{NO}} \right)^n}$$ $$\textrm{[PYeaR]}activity = \frac{\beta}{1+ \left( \frac{\textrm{NO}^*}{K_{d(\textrm{NsrR})}} \right)}$$ $$\frac{d[\textrm{mCP}]}{dt} = \frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} } - \gamma_m [\textrm{mCP}]$$ $$K=\frac{k_{transcription}[\textrm{PYeaR}]}{1 + \left(\frac{[\textrm{NsrR}]}{\left(1+\left[\frac{[\textrm{NO}]}{K_{NO}}\right]^n \right)}\right) k_{d(\textrm{NsrR})} }$$ $$\frac{d[\textrm{mCP}]}{dt} = K - \gamma_m [\textrm{mCP}]$$ $$[\textrm{mCP}]=\frac{K}{\gamma_m} - \frac{K}{\gamma_m}e^{-\gamma_mt}$$ $$[\textrm{CP}]=\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} e^{-\gamma_mt} - \left(\frac{K k_{translation}}{\gamma_p \gamma_m} + \frac{K k_{translation}}{{\gamma_m}^2-\gamma_p \gamma_m} \right)e^{-\gamma_pt} $$
| Parameter | Description | Value |
|---|---|---|
| $[\textrm{PYeaR}]$ | Concentration of PYeaR | 1 copy/cell |
| $k_{transcription}$ | Rate of chromoprotein mRNA synthesis | 0.167$\textrm{min}^{-1}$ |
| $k_{translation}$ | Rate of chromoprotein synthesis | $0.0011\textrm{min}^{-1}$ |
| $\gamma_m$ | mRNA degradation rate | $0.19\textrm{min}^{-1}$ |
| $\gamma_p$ | Chromoprotein degradation rate | $0.18\textrm{min}^{-1}$ |
| $K_{\textrm{NO}}$ | Dissociation constant of NO | $0.12\textrm{min}^{-1}$ |
| $K_{\textrm{d(NsrR)}}$ | Dissociation constant of NsrR | 0.035 mM |
| n | Cooperativity of NO binding to NsrR | 2 |
| Header Cell 1 | Header Cell 2 | Header Cell 3 |
|---|---|---|
| Row 1 Cell 1 | Row 1 Cell 2 | |
| Row 2 Cell 1 | Row 2 Cell 2 | |
| Row 3 Cell 1 | Row 3 Cell 2 | |
| Row 4 Cell 1 | Row 4 Cell 2 |


 Institute of Integrative Biosciences,
Institute of Integrative Biosciences,

 +92 91 5860291
+92 91 5860291



