Zigapusnik (Talk | contribs) |
Zigapusnik (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| − | + | {{Slovenia}} | |
| − | + | <html> | |
| − | + | <head> | |
| − | + | <title>Home</title> | |
| − | + | <link rel="stylesheet" | |
| − | + | href="//2016.igem.org/Team:Slovenia/libraries/semantic-min-css?action=raw&ctype=text/css"> | |
| − | + | <script type="text/javascript" | |
| − | + | src="//2016.igem.org/Team:Slovenia/libraries/semantic-min-js?action=raw&ctype=text/javascript"></script> | |
| − | + | <link rel="stylesheet" type="text/css" href="//2016.igem.org/Team:Slovenia/libraries/custom-css?action=raw&ctype=text/css"> | |
| − | + | <script type="text/javascript" src="//2016.igem.org/Team:Slovenia/libraries/custom-js?action=raw&ctype=text/javascript"></script> | |
| − | + | <script type="text/javascript" | |
| − | + | src="//2016.igem.org/Team:Slovenia/libraries/zitator-js?action=raw&ctype=text/javascript"></script> | |
| − | + | <script type="text/javascript" | |
| − | + | src="https://2016.igem.org/Team:Slovenia/libraries/bibtexparse-js?action=raw&ctype=text/javascript"></script> | |
| − | + | <!-- MathJax (LaTeX for the web) --> | |
| − | + | <script type="text/x-mathjax-config"> | |
| − | + | MathJax.Hub.Config({ | |
| − | + | tex2jax: { | |
| − | + | inlineMath: [['$', '$'], ['\\(', '\\)']] | |
| − | + | }, | |
| − | + | jax: ["input/TeX","output/SVG"], | |
| − | + | TeX: { | |
| − | + | extensions: ["mhchem.js"], | |
| − | + | equationNumbers: {autoNumber: "all" }, | |
| − | + | Macros: { | |
| − | + | goodbreak: '\\mmlToken{mo}[linebreak="goodbreak"]{}', | |
| − | + | badbreak: ['\\mmlToken{mo}[linebreak="badbreak"]{#1}',1], | |
| − | + | nobreak: ['\\mmlToken{mo}[linebreak="nobreak"]{#1}',1], | |
| − | + | invisibletimes: ['\\mmlToken{mo}{\u2062}'] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
} | } | ||
| − | } | + | }, |
| − | + | CommonHTML: { linebreaks: { automatic: true, width: "75% container" } }, | |
| − | + | "HTML-CSS": { linebreaks: { automatic: true, width: "68% container" }}, | |
| − | + | SVG: { linebreaks: { automatic: true, width: "200% container" }} | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
}); | }); | ||
| − | + | </script> | |
| − | + | <script type="text/javascript" async | |
| − | + | src="//2016.igem.org/common/MathJax-2.5-latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"> | |
| − | + | </script> | |
| − | + | </head> | |
| − | + | <body> | |
| − | + | <style> | |
| − | + | .container { | |
| − | + | position: relative; | |
| + | } | ||
| + | .module1 { | ||
| + | position: absolute; | ||
| + | top: 220px; | ||
| + | left: 10px; | ||
| + | } | ||
| + | .module2 { | ||
| + | position: absolute; | ||
| + | bottom: 90px; | ||
| + | left: 220px; | ||
| + | } | ||
| + | .module3 { | ||
| + | position: absolute; | ||
| + | top: 180px; | ||
| + | right: 350px; | ||
| + | } | ||
| + | .module4 { | ||
| + | position: absolute; | ||
| + | bottom: 15px; | ||
| + | right: 380px; | ||
| + | } | ||
| + | |||
| + | /* Popup container - can be anything you want */ | ||
| + | .popup { | ||
| + | display: inline-block; | ||
| + | cursor: pointer; | ||
| + | -webkit-user-select: none; | ||
| + | -moz-user-select: none; | ||
| + | -ms-user-select: none; | ||
| + | user-select: none; | ||
| + | padding: 10px 10px; | ||
| + | } | ||
| − | + | /* The actual popup */ | |
| − | + | .popup .popuptext { | |
| − | + | padding: 10px 10px; | |
| − | + | visibility: hidden; | |
| − | + | width: 350px; | |
| − | + | background-color: #555; | |
| − | + | color: #fff; | |
| − | + | text-align: center; | |
| − | + | border-radius: 6px; | |
| − | + | position: absolute; | |
| − | + | z-index: 100; | |
| − | + | font-size: 14px; | |
| − | } | + | line-height: 20px; |
| + | } | ||
| − | + | /* Toggle this class - hide and show the popup */ | |
| − | + | .popup .show { | |
| − | + | visibility: visible; | |
| + | -webkit-animation: fadeIn 1s; | ||
| + | animation: fadeIn 1s; | ||
| + | } | ||
| − | + | /* Add animation (fade in the popup) */ | |
| + | @-webkit-keyframes fadeIn { | ||
| + | from {opacity: 0;} | ||
| + | to {opacity: 1;} | ||
| + | } | ||
| − | + | @keyframes fadeIn { | |
| − | + | from {opacity: 0;} | |
| − | + | to {opacity:1 ;} | |
| − | + | } | |
| − | + | .container img{ | |
| − | + | display: block; | |
| − | + | margin-left: auto; | |
| − | + | margin-right: auto; | |
| + | } | ||
| + | |||
| − | + | .corners { | |
| − | + | border-radius: 15px; | |
| − | + | border: 2px solid #000000; | |
| − | + | padding: 15px; | |
| − | + | margin-left:15px; | |
| − | + | margin-bottom:15px; | |
| − | + | } | |
| − | + | ||
| − | + | </style> | |
| − | + | <script> | |
| − | + | $( document ).ready( | |
| − | + | function() { | |
| − | + | var getImgs = function() { | |
| − | + | var imgs = []; | |
| − | + | $('img').each(function() { | |
| − | + | console.log("image detected"); | |
| − | + | var data = $(this).data('alt'); | |
| − | + | imgs.push(data); | |
| − | + | }); | |
| − | + | return imgs; | |
| − | $( | + | } |
| − | + | var imgs = getImgs(); | |
| − | + | //preload images | |
| − | + | var image = []; | |
| − | + | $.each(imgs, function(index) { | |
| − | + | console.log("loading resource"); | |
| − | + | image[index] = new Image(); | |
| − | + | image[index].src = imgs[index]; | |
| − | + | }); | |
| − | + | } | |
| − | + | ); | |
| − | + | function loadImage(sourceImage, popupname) { | |
| − | + | console.log(sourceImage); | |
| − | + | var image = document.getElementById("projectScheme"); | |
| − | + | image.src = sourceImage; | |
| − | + | ||
| − | + | //var rect = image.getBoundingClientRect(); | |
| − | + | //console.log(rect.top, rect.right, rect.bottom, rect.left); | |
| − | + | ||
| − | + | var popup = document.getElementById(popupname); | |
| − | + | //popup.style.top = ((rect.bottom - rect.top)/2).toString().concat("px"); | |
| − | + | //popup.style.left = ((rect.right - rect.left)/2).toString().concat("px"); | |
| − | + | //console.log((rect.right - rect.left)/2, (rect.bottom - rect.top)/2); | |
| − | + | popup.classList.toggle('show'); | |
| − | + | } | |
| − | + | </script> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <div id="example"> | |
| − | + | <div class="pusher"> | |
| − | + | <div class="full height"> | |
| − | + | <div class="banana"> | |
| − | + | <a href = "//2016.igem.org/Team:Slovenia"> | |
| + | <img class="ui medium sticky image" src="//2016.igem.org/wiki/images/d/d1/T--Slovenia--logo.png"> | ||
| + | </a> | ||
| + | <div class="ui vertical sticky text menu"> | ||
| + | <a class="item" href="#intro" style="margin-left: 10%"> | ||
| + | <i class="selected radio icon"></i> | ||
| + | <b>Project</b> | ||
| + | </a> | ||
| + | <a class="item" href="#achievements" style="margin-left: 10%"> | ||
| + | <i class="selected radio icon"></i> | ||
| + | <b>Achievements</b> | ||
| + | </a> | ||
| + | <a class="item" href="#requirements" style="margin-left: 10%"> | ||
| + | <i class="selected radio icon"></i> | ||
| + | <b>Medal requirements</b> | ||
| + | </a> | ||
| + | <a class="item" href="//2016.igem.org/Team:Slovenia/Idea/Challenge"> | ||
| + | <i class="chevron circle right icon"></i> | ||
| + | <b>Idea</b> | ||
| + | </a> | ||
| + | <span> | ||
| + | <br /> | ||
| + | </span> | ||
| + | <img class = "ui large circular image" src="//2016.igem.org/wiki/images/c/cb/T--Slovenia--igemLogo.gif"> | ||
| + | <p style="font-size:11px;"> | ||
| + | The iGEM symbol was drawn with a glass rod letter by letter on engineered human cells and imaged by a camera. | ||
| + | Cells were transfected with constructs coding for the bacterial ion channel MscS, gas vesicles (GvpA and GvpC) and a Ca-dependent | ||
| + | cyclic split luciferase reporter. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | <div class="article" id="context"> | ||
| + | <!-- menu goes here --> | ||
| + | <!-- content goes here --> | ||
| + | <div class="main ui citing justified container"> | ||
| + | <h1 class="ui centered dividing header"><span id="intro" class = "section"> </span>Sonicell</h1> | ||
| + | <div class = "ui segment"> | ||
| + | <p>Project Sonicell introduces exciting foundational advances to synthetic biology aimed to enable rapid cellular response to a combination of external stimuli such as sound, light or chemical compounds. This system is composed of a module for enhanced sensitivity of cells to ultrasound or other mechanical stimuli sensed by a calcium-dependent reporter, and a module for integration of a combination of several input signals into a signaling pathway based on the collection of orthogonal proteases. Finally, the proteases were designed to cleave an endoplasmic reticulum retention signal from target proteins, which results in a secretion of premade proteins.</p> | ||
| + | <div> | ||
| + | <div class = "container"> | ||
| + | <img width = "100%" onload="relativeCoordinates(this);" style = "border-radius: 15px;" src="//2016.igem.org/wiki/images/c/c2/T--Slovenia--Main-scheme0.png" alt="project scheme" usemap="#projectmap" id = "projectScheme"></img> | ||
| + | <map name="projectmap"> | ||
| + | <area id="area1" class="popup" shape="poly" coords="" alt="module1" onmouseover="loadImage('//2016.igem.org/wiki/images/9/92/T--Slovenia--Main-scheme1.png', 'module1')" onmouseout="loadImage('//2016.igem.org/wiki/images/c/c2/T--Slovenia--Main-scheme0.png', 'module1')" href="//2016.igem.org/Team:Slovenia/Mechanosensing/Overview"></area> | ||
| + | <area id="area2" class="popup" shape="poly" coords="" alt="module1" onmouseover="loadImage('//2016.igem.org/wiki/images/5/54/T--Slovenia--Main-scheme3.png', 'module2')" onmouseout="loadImage('//2016.igem.org/wiki/images/c/c2/T--Slovenia--Main-scheme0.png', 'module2')" href="//2016.igem.org/Team:Slovenia/Protease_signaling/Logic"></area> | ||
| + | <area id="area3" class="popup" shape="poly" coords="" alt="module1" onmouseover="loadImage('//2016.igem.org/wiki/images/f/f6/T--Slovenia--Main-scheme2.png', 'module3')" onmouseout="loadImage('//2016.igem.org/wiki/images/c/c2/T--Slovenia--Main-scheme0.png', 'module3')" href="//2016.igem.org/Team:Slovenia/Protease_signaling/Overview"></area> | ||
| + | <area id="area4" class="popup" shape="poly" coords="" alt="module1" onmouseover="loadImage('//2016.igem.org/wiki/images/1/12/T--Slovenia--Main-scheme4.png', 'module4')" onmouseout="loadImage('//2016.igem.org/wiki/images/c/c2/T--Slovenia--Main-scheme0.png', 'module4')" href="//2016.igem.org/Team:Slovenia/Implementation/ProteaseInducible_secretion"></area> | ||
| + | </map> | ||
| + | <div class="popup module1"> | ||
| + | <span class="popuptext" id="module1"><b>Enhanced mechanosensing:</b><br /> Sensitivity of mammalian cells to ultrasound or other mechanical stimuli was enhanced by the introduction of mechanosensitive ion channels and/or by the expression of protein gas vesicles from bacteria. Influx of calcium through channels is sensed by formation of a complex between calmodulin and M13 peptide that can result in a rapid light emission by cells (used for cell painting) or reconstitution of a split protease.</span> | ||
| + | </div> | ||
| + | <div class="popup module2"> | ||
| + | <span class="popuptext" id="module2"><b>Protease based signaling <br />and information processing:</b><br /> Combinations of proteolytic activities against specific targets resulted in activation of a reporter or another protease, which forms the basis for the design of a new type of rapid signaling pathways and construction of logic functions.</span> | ||
| + | </div> | ||
| + | <div class="popup module3"> | ||
| + | <span class="popuptext" id="module3"><b>Orthogonal site-specific proteases: </b><br /> A collection of orthogonal site-specific proteases that recognize different targets was prepared as split proteins, whose activity against selected targets can be induced by stimulation with an external signal such as light or chemicals.</span> | ||
| + | </div> | ||
| + | <div class="popup module4"> | ||
| + | <span class="popuptext" id="module4"> <b>Protease-triggered rapid secretion <br /> of therapeutic proteins:</b> <br />A rapid cellular response by secretion of a protein is triggered by the proteolytic cleavage of an endoplasmic reticulum retention peptide. After the cleavage the cargo protein is moved from the ER, and secreted as therapeutic protein.</span> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class= "ui segment"> | ||
| + | <h4>Abstract for experts</h4> | ||
| + | <p>Synthetic biology opens exciting perspectives to control cells, for applications ranging from industrial processes to cell-based therapy. However, the large majority of designed cellular circuits are based on the transcriptional regulation, which may be too slow for many therapeutic or diagnostic applications, for example delivery of insulin or detection of a metabolite. Several medical doctors and researchers that we consulted stressed that a fast but controllable response is high on their wish list of expectations from synthetic biology. Additionally, noninvasive stimulation of selected tissues in the organism would also be highly desirable. While light is extremely useful as a rapid, spatially-restricted input signal, it cannot penetrate deep into the tissue. On the other hand, ultrasound combines several advantages of light with the added ability to penetrate tissue.</p> | ||
| + | <p>In our project we enhanced the sensitivity of mammalian cells to ultrasound or other mechanical stimulus by introduction of bacterial or engineered mammalian mechanosensors. Additionally, the response to ultrasound and touch was strongly increased by expression of the two components of bacterial gas vesicles, GvpA and GvpC. Mechanosensing was detected by the calcium-induced calmodulin-M13 complex reconstituting split cyclic luciferase, highly applicable for the emerging field of mechanogenetics. This enabled us to draw on cells using touch, where we engaged in collaboration with an artist.</p> | ||
| + | <p>For the rapid response of cells to multiple stimuli we designed proteolysis-based signaling pathways. For this purpose four orthogonal split proteases were generated, each recognizing its own motif of seven amino acid residues. Based on cleavage of coiled-coil dimerizing domains we demonstrated the ability to implement proteolysis-based signal pathways and logic functions in mammalian cells. Based on the cleavage of an ER retention peptide by a protease, input signals lead to protein secretion without the slow step of induced protein synthesis.</p> | ||
| + | <p>We believe that this project introduced several foundational advances that could be very useful to synthetic biology far beyond iGEM and for the benefit of humanity for therapy, diagnostics and potentially many other advanced applications.</p> | ||
| + | </div> | ||
| + | <div class = "ui segment"> | ||
| + | <h4>Abstract in plain English</h4> | ||
| + | <p> | ||
| + | Synthetic biology aims to control cells so they can obey our commands and do what we want, for example to produce drugs when needed. In our project we made cells respond to ultrasound or touch. When we touch the cells they light up, which can be recorded on a camera. Ideally we want cells to respond to our commands as fast as possible, because sometimes we can’t wait an hour before the cells produce the medicine and release it. That is why we gave cells a novel mechanism of processing information. | ||
| + | We achieved this by combining several enzymes that recognize very specific parts of proteins and cut them, which changes their function. This allowed us to combine different signals, like sound, touch, light or chemicals, to obtain the desired cell response. The new enzymes can also cut the anchor with which medicines are attached to cells after the cells make them. Among many possible uses of our inventions, we can imagine activating cells in the brain by ultrasound, which means that we don’t need to use | ||
| + | surgery to help people with Parkinson’s disease, or can trigger fast production of insulin in the body, to help people with diabetes. | ||
| + | </p> | ||
| + | </div> | ||
| + | <div> | ||
| + | <h1 class="ui centered dividing header"><span id="achievements" class = "section"> </span>Achievements</h1> | ||
| + | <div class = "ui segment"> | ||
| + | <div class = "corners" style="float:right;"> | ||
| + | <p><img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"> new at science</p> | ||
| + | <p><img src="//2016.igem.org/wiki/images/0/0d/T--Slovenia--igemLogoSmall.png" alt="newAtiGEM" width="30" height="30" style = "display: inline;"> new at iGEM</p> | ||
| + | </div> | ||
| + | <ul> | ||
| + | <li>Mammalian cell sensitivity to ultrasound and mechanical stimuli was increased by ectopic expression of bacterial or human cation permeable channels and functional reconstitution of bacterial protein gas vesicles from two protein components (GvpA and GvpC) <img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"></li> | ||
| + | <li>A custom-made ultrasound generator device was used to stimulate mammalian cells <img src="//2016.igem.org/wiki/images/0/0d/T--Slovenia--igemLogoSmall.png" alt="newAtiGEM" width="30" height="30" style = "display: inline;"></li> | ||
| + | <li>A mechano-sensory luciferase reporter sensitive to an influx of free calcium ions was introduced into mammalian cells, which enabled rapid light emission of mammalian cells in response to mechanical stimuli and enabled painting on cells by touch with exciting potentials for other applications <img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"></li> | ||
| + | <li>A circular proteolysis-activated luciferase reporter was experimentally verified and introduced into the iGEM collection <img src="//2016.igem.org/wiki/images/0/0d/T--Slovenia--igemLogoSmall.png" alt="newAtiGEM" width="30" height="30" style = "display: inline;"></li> | ||
| + | <li>A set of four different orthogonal site-specific proteases was designed and tested as split proteins in mammalian cells <img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"></li> | ||
| + | <li>New orthogonal protease-based signaling pathways and information processing platform was designed and several logic functions based on the combination of multiple input signals were tested experimentally <img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"></li> | ||
| + | <li>Proteolysis of the ER retention signal was introduced as the trigger for the fast release of proteins from cells aimed to enable fast therapeutic responses such as required for the release of peptide hormones, neuroactive peptides etc. <img src="//2016.igem.org/wiki/images/d/dc/T--Slovenia--starSmall.png" alt="newAtiGEM" width="28" height="28" style = "display: inline;"></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div> | ||
| + | <h1 class="ui centered dividing header"><span id="requirements" class = "section"> </span>Medal requirements</h1> | ||
| + | <div class = "ui segment"> | ||
| + | <h4>Gold:</h4> | ||
| + | <h4>Silver:</h4> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
Revision as of 13:28, 17 October 2016
Sonicell
Project Sonicell introduces exciting foundational advances to synthetic biology aimed to enable rapid cellular response to a combination of external stimuli such as sound, light or chemical compounds. This system is composed of a module for enhanced sensitivity of cells to ultrasound or other mechanical stimuli sensed by a calcium-dependent reporter, and a module for integration of a combination of several input signals into a signaling pathway based on the collection of orthogonal proteases. Finally, the proteases were designed to cleave an endoplasmic reticulum retention signal from target proteins, which results in a secretion of premade proteins.
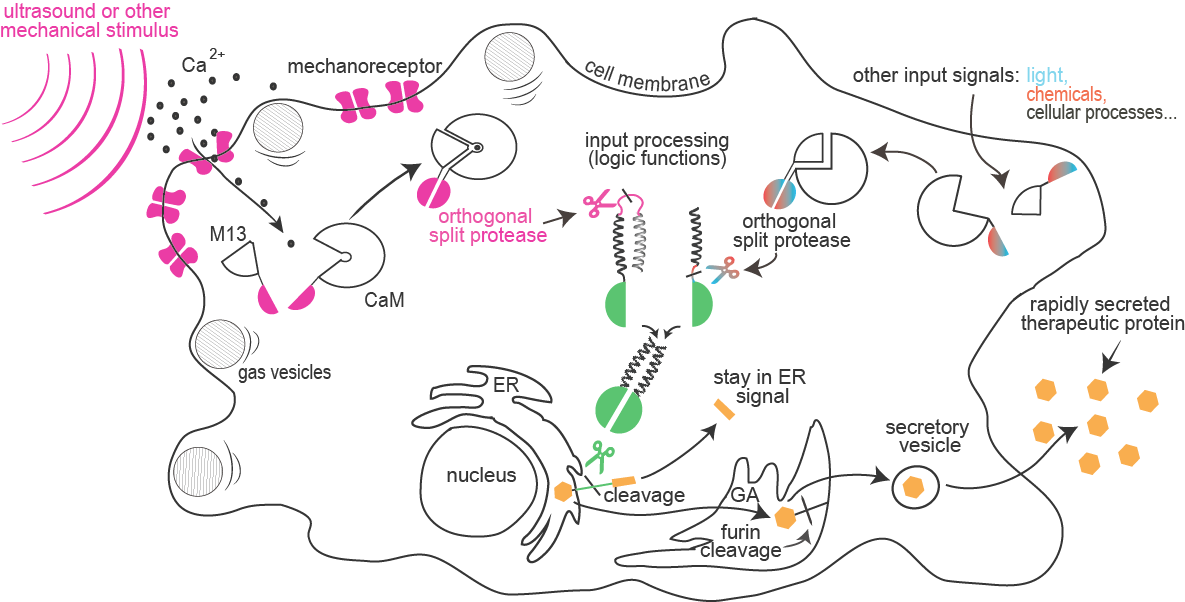
Sensitivity of mammalian cells to ultrasound or other mechanical stimuli was enhanced by the introduction of mechanosensitive ion channels and/or by the expression of protein gas vesicles from bacteria. Influx of calcium through channels is sensed by formation of a complex between calmodulin and M13 peptide that can result in a rapid light emission by cells (used for cell painting) or reconstitution of a split protease.
and information processing:
Combinations of proteolytic activities against specific targets resulted in activation of a reporter or another protease, which forms the basis for the design of a new type of rapid signaling pathways and construction of logic functions.
A collection of orthogonal site-specific proteases that recognize different targets was prepared as split proteins, whose activity against selected targets can be induced by stimulation with an external signal such as light or chemicals.
of therapeutic proteins:
A rapid cellular response by secretion of a protein is triggered by the proteolytic cleavage of an endoplasmic reticulum retention peptide. After the cleavage the cargo protein is moved from the ER, and secreted as therapeutic protein.
Abstract for experts
Synthetic biology opens exciting perspectives to control cells, for applications ranging from industrial processes to cell-based therapy. However, the large majority of designed cellular circuits are based on the transcriptional regulation, which may be too slow for many therapeutic or diagnostic applications, for example delivery of insulin or detection of a metabolite. Several medical doctors and researchers that we consulted stressed that a fast but controllable response is high on their wish list of expectations from synthetic biology. Additionally, noninvasive stimulation of selected tissues in the organism would also be highly desirable. While light is extremely useful as a rapid, spatially-restricted input signal, it cannot penetrate deep into the tissue. On the other hand, ultrasound combines several advantages of light with the added ability to penetrate tissue.
In our project we enhanced the sensitivity of mammalian cells to ultrasound or other mechanical stimulus by introduction of bacterial or engineered mammalian mechanosensors. Additionally, the response to ultrasound and touch was strongly increased by expression of the two components of bacterial gas vesicles, GvpA and GvpC. Mechanosensing was detected by the calcium-induced calmodulin-M13 complex reconstituting split cyclic luciferase, highly applicable for the emerging field of mechanogenetics. This enabled us to draw on cells using touch, where we engaged in collaboration with an artist.
For the rapid response of cells to multiple stimuli we designed proteolysis-based signaling pathways. For this purpose four orthogonal split proteases were generated, each recognizing its own motif of seven amino acid residues. Based on cleavage of coiled-coil dimerizing domains we demonstrated the ability to implement proteolysis-based signal pathways and logic functions in mammalian cells. Based on the cleavage of an ER retention peptide by a protease, input signals lead to protein secretion without the slow step of induced protein synthesis.
We believe that this project introduced several foundational advances that could be very useful to synthetic biology far beyond iGEM and for the benefit of humanity for therapy, diagnostics and potentially many other advanced applications.
Abstract in plain English
Synthetic biology aims to control cells so they can obey our commands and do what we want, for example to produce drugs when needed. In our project we made cells respond to ultrasound or touch. When we touch the cells they light up, which can be recorded on a camera. Ideally we want cells to respond to our commands as fast as possible, because sometimes we can’t wait an hour before the cells produce the medicine and release it. That is why we gave cells a novel mechanism of processing information. We achieved this by combining several enzymes that recognize very specific parts of proteins and cut them, which changes their function. This allowed us to combine different signals, like sound, touch, light or chemicals, to obtain the desired cell response. The new enzymes can also cut the anchor with which medicines are attached to cells after the cells make them. Among many possible uses of our inventions, we can imagine activating cells in the brain by ultrasound, which means that we don’t need to use surgery to help people with Parkinson’s disease, or can trigger fast production of insulin in the body, to help people with diabetes.
Achievements
![]() new at science
new at science
![]() new at iGEM
new at iGEM
- Mammalian cell sensitivity to ultrasound and mechanical stimuli was increased by ectopic expression of bacterial or human cation permeable channels and functional reconstitution of bacterial protein gas vesicles from two protein components (GvpA and GvpC)

- A custom-made ultrasound generator device was used to stimulate mammalian cells

- A mechano-sensory luciferase reporter sensitive to an influx of free calcium ions was introduced into mammalian cells, which enabled rapid light emission of mammalian cells in response to mechanical stimuli and enabled painting on cells by touch with exciting potentials for other applications

- A circular proteolysis-activated luciferase reporter was experimentally verified and introduced into the iGEM collection

- A set of four different orthogonal site-specific proteases was designed and tested as split proteins in mammalian cells

- New orthogonal protease-based signaling pathways and information processing platform was designed and several logic functions based on the combination of multiple input signals were tested experimentally

- Proteolysis of the ER retention signal was introduced as the trigger for the fast release of proteins from cells aimed to enable fast therapeutic responses such as required for the release of peptide hormones, neuroactive peptides etc.




