| Line 38: | Line 38: | ||
<div class="labbook"> | <div class="labbook"> | ||
</div> | </div> | ||
| + | |||
| + | =Samples= | ||
| + | <div class="week" id="WWeek_0"> | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | === Transformation of ''E. coli'' XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of Phytochrome B for protein fusion. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 2 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 1 ml LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker. | ||
| + | |||
| + | * 100 µl of the cell suspension was plated on one chloramphenicol plate. | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded. | ||
| + | |||
| + | * The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate. | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario''' | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Sequencing of RFP-Generator (RFC25, pSB1C3) === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of RFP-Generator (RFC25, pSB1C3) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) | ||
| + | </div> | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | |||
| + | === Picking of of ''E. coli'' XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Picking of of ''E. coli'' XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates. | ||
| + | |||
| + | * Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x). | ||
| + | |||
| + | * 4 colonies were picked. | ||
| + | |||
| + | * These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5) === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5). | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P4 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P4 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P5 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P5 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Incubation for 90 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp ladder DNA ladder | ||
| + | |'''P4''' | ||
| + | |'''P5''' | ||
| + | |- | ||
| + | | | ||
| + | |'''Mutation successful''' | ||
| + | |'''Mutation successful!''' | ||
| + | |} | ||
| + | |||
| + | * Parts are compliant and do not contain RFC25 forbidden restriction sites. | ||
| + | |||
| + | |||
| + | [[File:TUM13_20130423_RFP_Generator_RFC25_AgeI_NgoMIV.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2 === | ||
| + | |||
| + | '''Investigator: Jeff, Rosario, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). | ||
| + | |||
| + | The different vectors we sequenced received the following barcodes: | ||
| + | |||
| + | - ADH in pTUM100: FR01002265 | ||
| + | |||
| + | - TEF1 in pTUM100: FR01002266 | ||
| + | |||
| + | - TEF2 in pTUM100: FR01002266 | ||
| + | |||
| + | - GAL in pTUM100: FR01002268 | ||
| + | |||
| + | |||
| + | Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry. | ||
| + | </div> | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | |||
| + | === Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| + | |||
| + | '''Investigator: Jeff, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3). | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="safety_mechanism"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10 === | ||
| + | |||
| + | '''Investigator: Jeff, Florian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Batch for analytical digestion for P7 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P7 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P8 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P8 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P9 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P9 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion for P10 with NgoMIV+AgeI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA P10 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |NgoMIV (10 U/µl) | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI-HF (20 U/µl) | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Incubation for 90 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp ladder DNA ladder | ||
| + | |'''P7''' | ||
| + | |'''P8''' | ||
| + | |'''P9''' | ||
| + | |'''P10''' | ||
| + | |- | ||
| + | | | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |'''Part is correct''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[File:TUM13_20130424_PhytochromeB_RFC25_AgeI_NgoMIV.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="effectors"> | ||
| + | |||
| + | === Transformation of ''E. coli'' XL1 blue with === | ||
| + | |||
| + | '''Investigator: ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 1, May 16th - May 22nd= | ||
| + | <div class="week" id="WWeek_1"> | ||
| + | |||
| + | =='''Monday, May 16th'''== | ||
| + | <div class="general"> | ||
| + | === Streptavidin plasmids control === | ||
| + | |||
| + | '''Investigator: JB, LK, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) (4 clones each of pSA1, pSAm1 in pASK75) | ||
| + | * analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF), 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O | ||
| + | * 5 µl on 1% agarose gel electrophoresis of digestion | ||
| + | |||
| + | '''Results:''' successful cloning verified, stored at -20 °C | ||
| + | |||
| + | * 1. Lane: 5 µl Thermo Fisher, 1kb Ladder | ||
| + | |||
| + | * 2. to 9. Lane: 5 µl digestions of P6 to P13, band of SA (mut1) at about 300 bp, band of digested plasmid at about 3.000 bp | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Streptavidin expression_trafo BL21 === | ||
| + | |||
| + | '''Investigator: JB, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' expression of pSA1 and pSAm1 in E. Coli BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * transformation according to protocol of P6 and P10 in competent E. Coli BL21 | ||
| + | |||
| + | '''result:''' plates (LB Amp) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, May 17th'''== | ||
| + | <div class="general"> | ||
| + | === SDS Gel Analysis === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' SDS gel analysis of collagen 1/2, eGFP, fraction 30 of egg-precipitation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * mixing of 80 µl samples with 20 µl SDS buffer and heating at 95°C for 10 min. 1 d staining, 1 d unstaining | ||
| + | |||
| + | '''Results:''' successful cloning verified, stored at -20 °C | ||
| + | |||
| + | * 1. Lane: 8 µl Marker (Thermo Fisher #26610) | ||
| + | |||
| + | * 2. Lane: fraction 30 (IEC), 3 µl, band at 35 kDa, Avidin expected at 16 kDa | ||
| + | |||
| + | * 3. Lane: eGFP, 12 µl, band at 27 kDa eGFP expected at 27 kDa, many impurities | ||
| + | |||
| + | * 4. Lane: Collagen 1, 12 µl, no sharp band | ||
| + | |||
| + | * 5. Lane: Collagen 1, 12 µl, no sharp band | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Minipreps pSb1C3-AviTag, -A3C5, pASK75-(SA1), -(SAm1) === | ||
| + | |||
| + | '''Investigator: CR, CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | |||
| + | * analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF) for pASK plasmids and 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O - 5 µl on 1% agarose gel electrophoresis of digestion | ||
| + | |||
| + | '''result:''' : successful cloning verified for pASK plasmids, repetition of pSb1C3 plasmids, stored at -20 °C | ||
| + | |||
| + | * 1. Lane: 5 µl Thermo Fisher, 1 kb Ladder | ||
| + | |||
| + | * 2. Lane: 5 µl digestion of pSb1C3-AviTag | ||
| + | |||
| + | * 3. Lane: 5 µl digestion of pSb1C3-A3C5 | ||
| + | |||
| + | * 5. Lane: 5 µl digestion of pASK75(SA1), EB elution | ||
| + | |||
| + | * 6. Lane: 5 µl digestion of pASK75(SAmut1), EB elution | ||
| + | |||
| + | * 7. Lane: 5 µl digestion of pASK75(SAmut1), H2O elution | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of pre-culture with BL21 (pASK75 (SA1)) in LB-medium === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Preculture for streptavidin expression in TB-medium | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Add 50 µL ampicillin in 50 mL LB-medium | ||
| + | |||
| + | * Picking colonies from BL21 (pASK75 (SA1)) | ||
| + | |||
| + | * Inoculate LB-medium | ||
| + | |||
| + | * Incubate at 30°C over night | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, May 18th'''== | ||
| + | <div class="general"> | ||
| + | === Repetition of analytical gel of pSb1C3-AviTag, -A3C5 === | ||
| + | |||
| + | '''Investigator: CG, CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * analytic digestion with: 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 8,5 µl Plasmid-DNA | ||
| + | * 10 µl on 2% agarose gel electrophoresis of digestion | ||
| + | |||
| + | '''Results:''' | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Production of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Ampicillin (2 mL) was added to the Medium (1:1000) | ||
| + | * The pre-culture (50 mL) was poured into the Medium | ||
| + | * Culture was incubated at 37°C and 140 rpm until OD550 reached 0.5 | ||
| + | * To induce streptavidin expression anhydro-tetrazycline (200 µL) was added to the culture (1:10000) | ||
| + | * The culture was incubated at 37°C and 140 rpm for 4 hours | ||
| + | |||
| + | '''Results:''' | ||
| + | * Streptavidin expression by BL21 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium === | ||
| + | |||
| + | '''Investigator: CR, JB, JH ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of Streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4°C, 5000 rpm, 20 mins, F 4X1L rotor) | ||
| + | * The supernatant was cast away and the pellet was transferred into a beaker of sufficient size and resuspended in fridge-cooled Tris Buffer B (50 mM Tris/HCl (pH = 8.0), 1 mM EDTA) | ||
| + | * The solution was homogenized in the PANDA (ask supervisor) | ||
| + | * The resulting lysate was transferred into centrifuge tubes and spun down (4°C, 18,000 rpm, 10 mins, XX34-rotor). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight. | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Dialysis of eGFP === | ||
| + | |||
| + | '''Investigator: NA, JH, CR ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * eGFP was thawed on ice | ||
| + | * eGFP was then poured in a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in ice cold Tris/HCl 20 mM pH 8.0 | ||
| + | * The dialysis took place at 4°C over nightXX34-rotor). The supernatant was cast away and the pellet was | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MiniPrep of quickchanged pNGAL146-A2 === | ||
| + | |||
| + | '''Investigator: NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extraction of pNGAL146-A2 plasmid from XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Sequencing of P14, P15 & P19 === | ||
| + | |||
| + | '''Investigator: CR, NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P14, P15 & P19 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | * The different plasmids we prepared received the following barcodes: | ||
| + | * P14 : FR11326653 | ||
| + | * P15 : FR11326655 | ||
| + | * P19 (K4): FR11326654 | ||
| + | * P16 (K1): FR11326652 | ||
| + | * P17 (K2): FR11326651 | ||
| + | * P18 (K3): FR11326650 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Digestion of P16, P17, P18 & P19 with AgeI & HindIII + analytical gel === | ||
| + | |||
| + | '''Investigator: NA, JH, CR ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of success of quickchange | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * analytic digestion with: 0,25 µl HindIII (HF), 0,25 µl AgeI (HF), 1 µl SmartCut Buffer, 500 ng plasmid-DNA, fill up with ddH2O (Vtotal= 10µL) | ||
| + | * 10 µl on 2% agarose gel for electrophoresis | ||
| + | |||
| + | '''Results:''' No signal at 600 bp --> quickchange seems to be successful (waiting for sequencing) | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, May 19th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Re-Sequencing of P19=== | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Re-Sequencing of P19 | ||
| + | |||
| + | '''Procedure:'''Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | |||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | *P19 (K4): FR11326649 | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Cloning of A3C5 and Avi-Tag into pSB1C3=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Re-Trafo of pSB1C3 RFP for later on: digestion, dephosphorylation and cloning | ||
| + | |||
| + | '''Procedure:''' transformation according to protocol of P4 E. Coli XL1 | ||
| + | |||
| + | '''Result:''' plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Streptavidin refolding=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Refolding of denaturated Streptavidin | ||
| + | |||
| + | '''Procedure:''' After the pellet had almost completely dissolved in 6M GdmCl, the solution was spun down (4°C, 20 mins, 18,000 rpm). The supernatant was transferred carefully into a falcon tube and the pellet was cast away. Via a hydraulic pump (flow rate: 2x10 ml/min) the lysate was transferred Into 5L PBS 1x. Afterwards the pump was cleaned with technical isopropanol and ELGA water. The solution was stirred overnight at 4°C for refolding. | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Biotinylation of BSA=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Biotinylation of BSA | ||
| + | |||
| + | '''Procedure:''' A 100 µM (=6.8 mg/ml) solution of BSA (Albumin fraction V, pH=7, in the fridge in the central lab) was created (V=10 ml). 220 µl of a 100 mM Biotin stock were added. The mixture was stored overnight Iin the fridge (4°C). | ||
| + | |||
| + | '''Result:''' Hopefully biotinylated BSA mixture in the fridge (4°C). | ||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 2 (May 23rd - May 29th= | ||
| + | <div class="week" id="WWeek_2"> | ||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 3 (May 30th - June 5th= | ||
| + | <div class="week" id="WWeek_3"> | ||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 4 (June 6th - June 12th)= | ||
| + | <div class="week" id="WWeek_4"> | ||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 5 (June 13th - June 19th)= | ||
| + | <div class="week" id="WWeek_5"> | ||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 6 (June 20th - June 26th)= | ||
| + | <div class="week" id="WWeek_6"> | ||
| + | |||
| + | =='''Thursday, June 23rd'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001=== | ||
| + | |||
| + | '''Investigator: Jan, Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001 | ||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table class="wikitable" cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P80 | ||
| + | </td><td>432,7 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P81 | ||
| + | </td><td>294,8 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P82 | ||
| + | </td><td>450,5 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P83 | ||
| + | </td><td>479,0 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P84 | ||
| + | </td><td>108,0 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P85 | ||
| + | </td><td>356,0 | ||
| + | </td></tr><tr> | ||
| + | <td>P86 | ||
| + | </td><td>47,2 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
| + | =='''Friday, June 24th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
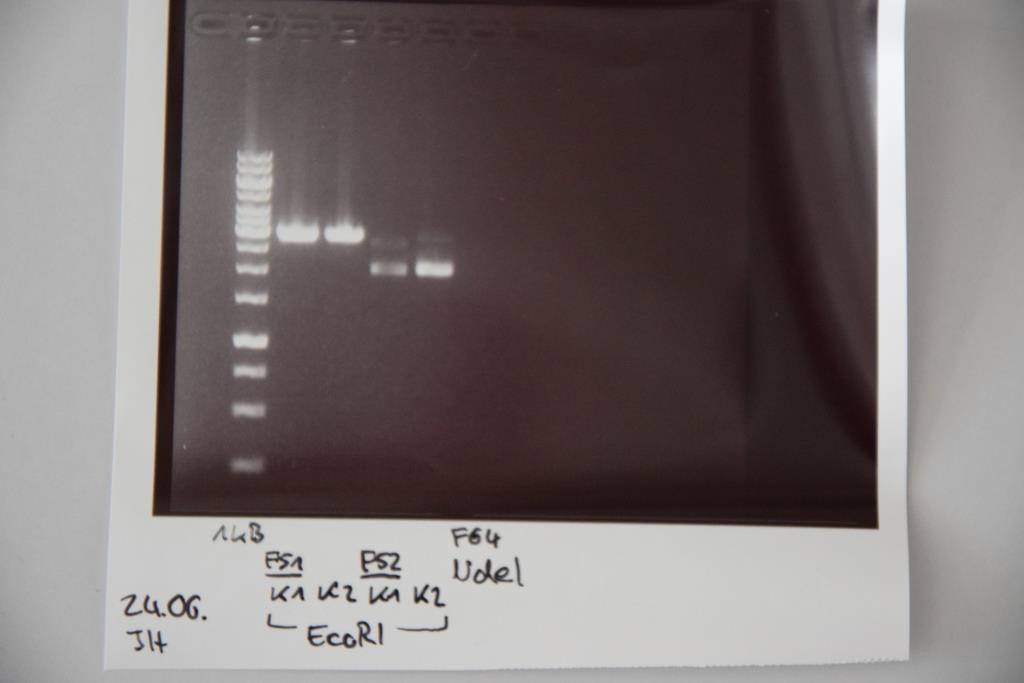
| + | === Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2) === | ||
| + | |||
| + | '''Investigator: Julian, Niklas, Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of P82/83 (EspP K1/2) and P84/85 (StrepTag K1/2). | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P82-P85 with EcoRI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.5/1.0 µl | ||
| + | |Plasmid DNA (-/P84) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl) | ||
| + | |- | ||
| + | |8/7.5 µl | ||
| + | |ddH2O (-/P84) | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_P82-85_EcoRI.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). Sequencing primer VF2 was used | ||
| + | |||
| + | The different vectors we sequenced received the following barcodes: | ||
| + | |||
| + | * mRuby3 in pSB1C3 (P80): FR11326590 | ||
| + | |||
| + | * EspP in pSB1C3 (P83): FR11326588 | ||
| + | |||
| + | * Streptag in pSB1C3 (P85): FR11326587 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of ''E. coli'' XL1 blue with F64 (quickchanged P3(pSAm1)) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | --> Quickchange did not work, do again, than new transformation! | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Gelextraction of F67(BirA(Digest. F59 [EcoRI; SpeI])), F68(mRuby(Digest. F60 [NgoMIV; SpeI])), F69(EGFR-TMD(Digest. F60 [NgoMIV; SpeI]), F70(pSB1C3(digest. P74 [NgoMIV; SpeI]) and F71(pSB1C3(digest. P74 [EcoRI; SpeI]) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Gelextraction was performed by manufacturers protocol (Qiagen). | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, June 25th'''== | ||
| + | <div class="Receptor"> | ||
| + | ===Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P87 | ||
| + | </td><td>81 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P88 | ||
| + | </td><td>34,5 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P89 | ||
| + | </td><td>86,3 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P90 | ||
| + | </td><td>108,5 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P91 | ||
| + | </td><td>417,4 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
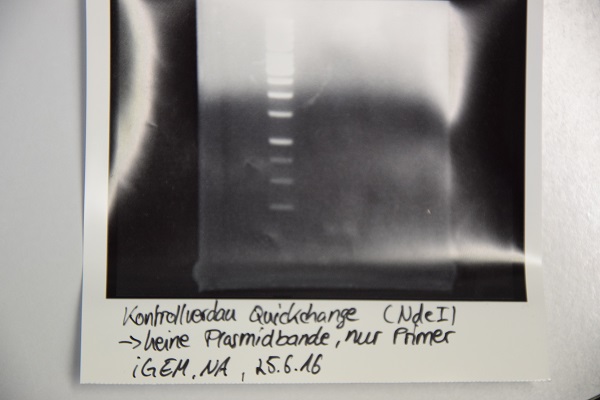
| + | === Analytical digestion and gelelectrophoresis of F64 (quickchanged P3) for verification of succesful quickchange === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Analytical digestion with NdeI and gelelectrophoresis. If quickchange worked there should be a band at about 3200 bp (only one restriction site left) | ||
| + | |||
| + | * Incubation over night at room temperature. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Quickchange_NA.JPG]] | ||
| + | |||
| + | Just a band showing a few bp (Primer), there is no plasmid band -> Quickchange did not work | ||
| + | </div> | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
| + | === Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3)and F44 + F30 (mRuby in pSB1C3) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 6x 4 ml LB+Cam media | ||
| + | |||
| + | * Each culture was inoculated with one colony | ||
| + | |||
| + | * Incubation at 37°C overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Sunday, June 26th'''== | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
| + | ===Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extracting F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) from E.coli XL-1-blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P92 | ||
| + | </td><td>162,9 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P93 | ||
| + | </td><td>447,9 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
| + | === Repetition of Quick-Change PCR of P3 (pASK + SAm1) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The QC-PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O21 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O22 | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 5 µl | ||
| + | </td><td>Pfu-Ultra-II reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td>Pfu-Ultra-II Polymerase | ||
| + | <tr> | ||
| + | <td>40,5 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | * digestion of PCR-Product with DpnI for 1h at 37°C | ||
| + | |||
| + | * now labeled P94 | ||
| + | </div> | ||
| + | |||
| + | === Transformation of ''E. coli'' XL1 blue with P94 (quickchanged P3(pSAm1)) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) for pASK (F72) and on chloramphenicol plates for P92 and P93 and incubated over night at 37 °C in the incubator. | ||
| + | |||
| + | <div class="Receptor"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75). | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P92 and P93 with EcoRI-HF and PstI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl) and PstI-HF (10 U/µl) for P92/93, NdeI (10 U/µl) for P94 | ||
| + | |- | ||
| + | |7.5/7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * 1. band (P92): no mRuby at 700bp visible, only empty vector | ||
| + | * 2. band (P93): empty vector and EGFR --> perfect | ||
| + | * 3. band (P94): no signal at all --> repetition of QC-PCR | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 7 (June 27th - July 3rd)= | ||
| + | <div class="week" id="WWeek_7"> | ||
| + | |||
| + | =='''Monday, June 27th'''== | ||
| + | === Sequencing of P67 (EGFR-Signalpeptid) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2)) | ||
| + | |||
| + | FR11326586 | ||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | |||
| + | <div> | ||
| + | === Repetition of Quick-Change PCR of P3 (pASK + SAm1) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The QC-PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O21 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O22 | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 5 µl | ||
| + | </td><td>Pfu-Ultra-II reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td>Pfu-Ultra-II Polymerase | ||
| + | <tr> | ||
| + | <td>40,5 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | * Digestion of PCR-Product with DpnI for 1h at 37°C. | ||
| + | |||
| + | * Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR). | ||
| + | </div> | ||
| + | |||
| + | === PCR of Genesynthesis 3 and 4 === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 2,5 µl | ||
| + | </td><td>Primer VF2 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 2,5 µl | ||
| + | </td><td>Primer VR2 | ||
| + | <tr> | ||
| + | <td> 1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 10 µl | ||
| + | </td><td> Q5 Polymerase reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td> Q5-Polymerase | ||
| + | <tr> | ||
| + | <td> 18 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | *Setup: iGEM_standard (Promega-cycler) | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>temperature</b> | ||
| + | </td><td><b>time</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 98°C | ||
| + | </td><td> 2min | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 98°C | ||
| + | </td><td> 10sec | ||
| + | <tr> | ||
| + | <td> 66°C | ||
| + | </td><td> 30sec | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 72°C | ||
| + | </td><td> 30sec | ||
| + | <tr> | ||
| + | <td> 72°C | ||
| + | </td><td> 2min | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 4°C | ||
| + | </td><td> hold | ||
| + | </td></tr></table> | ||
| + | |||
| + | * the batches were then purified using the Quiagen PCR-Purification Kit. | ||
| + | |||
| + | </div> | ||
| + | |||
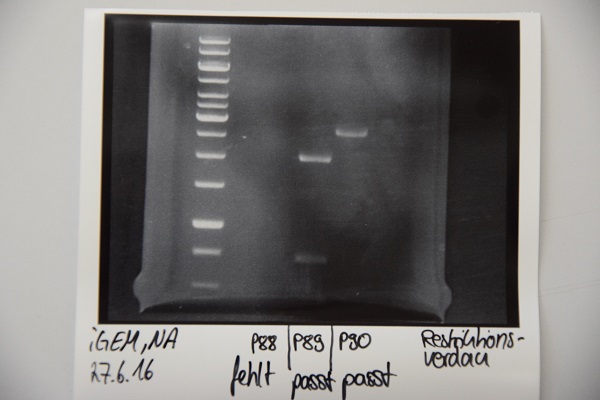
| + | === Analytical digestion and gelelectrophoresis of P88 , P89 and P90 === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | P88: EcoRI | ||
| + | |||
| + | P89: EcoRI and PstI | ||
| + | |||
| + | P90: EcoRI | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |5,8/2,3/1,8 µl | ||
| + | |Plasmid DNA (P88/P89/P90) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl)/ PstI | ||
| + | |- | ||
| + | |required amount for total volume of 10 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_P88-90_NA.JPG]] | ||
| + | |||
| + | === Ligation of F67 and F71, Transformation of ''E. coli'' XL1 blue afterwards === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of ''E. coli'' XL1 blue afterwards. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2,4 µl | ||
| + | |Vektor | ||
| + | |- | ||
| + | |7,6 µl | ||
| + | |Insert | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | * next step: analytic digestion of transformation was successful | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | |||
| + | === Digestion of PCR on genesynthesis 3 and 4, and pSB1C3 === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl each | ||
| + | |enzyme (SapI, HindIII, XbaI, AgeI) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |41 µl | ||
| + | |DNA (purified PCR-products of GSY3 and 4) | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | * Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of P80 , P78 and P85 === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: Analytical digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85(Strep-Tag)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | *P80 and P78: EcoRI and AgeI | ||
| + | |||
| + | *P85: EcoRI and NgoMIV | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |32 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |each enzyme(10 U/µl)/ PstI | ||
| + | |- | ||
| + | |50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_.JPG]] | ||
| + | |||
| + | === Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into ''E. coli'' XL1 blue === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of ''E. coli'' XL1 blue afterwards. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |4,3µl(for F75), 8,2µl (for F76) | ||
| + | |Vector | ||
| + | |- | ||
| + | |12,7µl (F75), 8,8 (F76) | ||
| + | |Insert | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Ligation was incubated at RT for 1,5h. | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 15 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator. | ||
| + | |||
| + | === Chemical biotinylation of BSA === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * BSA was chemically biotinylated with a 20x and 40x molar excess: | ||
| + | |||
| + | * 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85) | ||
| + | |||
| + | * dissolve BSA (10 mg/ml) | ||
| + | |||
| + | * Add biotin-NHS-ester: 20,5 mg for 40x molar excess | ||
| + | |||
| + | * reaction over night | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 28th'''== | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 29th'''== | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
Revision as of 17:07, 29 June 2016
Contents
- 1 Labjournal
- 2 Samples
- 2.1 Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
- 2.2 Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
- 2.3 Sequencing of RFP-Generator (RFC25, pSB1C3)
- 2.4 Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
- 2.5 Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
- 2.6 Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
- 2.7 Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
- 2.8 Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
- 2.9 Transformation of E. coli XL1 blue with
- 3 Week 1, May 16th - May 22nd
- 3.1 Monday, May 16th
- 3.2 Tuesday, May 17th
- 3.3 Wednesday, May 18th
- 3.3.1 Repetition of analytical gel of pSb1C3-AviTag, -A3C5
- 3.3.2 Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline
- 3.3.3 Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium
- 3.3.4 Dialysis of eGFP
- 3.3.5 MiniPrep of quickchanged pNGAL146-A2
- 3.3.6 Sequencing of P14, P15 & P19
- 3.3.7 Digestion of P16, P17, P18 & P19 with AgeI & HindIII + analytical gel
- 3.4 Thursday, May 19th
- 4 Week 2 (May 23rd - May 29th
- 5 Week 3 (May 30th - June 5th
- 6 Week 4 (June 6th - June 12th)
- 7 Week 5 (June 13th - June 19th)
- 8 Week 6 (June 20th - June 26th)
- 8.1 Thursday, June 23rd
- 8.2 Friday, June 24th
- 8.2.1 Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2)
- 8.2.2 Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
- 8.2.3 Transformation of E. coli XL1 blue with F64 (quickchanged P3(pSAm1))
- 8.2.4 Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3)
- 8.3 Saturday, June 25th
- 8.3.1 Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)
- 8.3.2 Analytical digestion and gelelectrophoresis of F64 (quickchanged P3) for verification of succesful quickchange
- 8.3.3 Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3)and F44 + F30 (mRuby in pSB1C3)
- 8.4 Sunday, June 26th
- 8.4.1 Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3)
- 8.4.2 Repetition of Quick-Change PCR of P3 (pASK + SAm1)
- 8.4.3 Transformation of E. coli XL1 blue with P94 (quickchanged P3(pSAm1))
- 8.4.4 Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75)
- 9 Week 7 (June 27th - July 3rd)
- 9.1 Monday, June 27th
- 9.1.1 Sequencing of P67 (EGFR-Signalpeptid)
- 9.1.2 Repetition of Quick-Change PCR of P3 (pASK + SAm1)
- 9.1.3 PCR of Genesynthesis 3 and 4
- 9.1.4 Analytical digestion and gelelectrophoresis of P88 , P89 and P90
- 9.1.5 Ligation of F67 and F71, Transformation of E. coli XL1 blue afterwards
- 9.1.6 Digestion of PCR on genesynthesis 3 and 4, and pSB1C3
- 9.1.7 Analytical digestion and gelelectrophoresis of P80 , P78 and P85
- 9.1.8 Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into E. coli XL1 blue
- 9.1.9 Chemical biotinylation of BSA
- 9.2 Tuesday, June 28th
- 9.3 Wednesday, June 29th
- 9.1 Monday, June 27th
Labjournal
Samples
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with
Investigator:
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
Week 1, May 16th - May 22nd
Monday, May 16th
Streptavidin plasmids control
Investigator: JB, LK, JH
Aim of the experiment: Verification of cloning
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) (4 clones each of pSA1, pSAm1 in pASK75)
- analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF), 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O
- 5 µl on 1% agarose gel electrophoresis of digestion
Results: successful cloning verified, stored at -20 °C
- 1. Lane: 5 µl Thermo Fisher, 1kb Ladder
- 2. to 9. Lane: 5 µl digestions of P6 to P13, band of SA (mut1) at about 300 bp, band of digested plasmid at about 3.000 bp
Streptavidin expression_trafo BL21
Investigator: JB, JH
Aim of the experiment: expression of pSA1 and pSAm1 in E. Coli BL21
Procedure:
- transformation according to protocol of P6 and P10 in competent E. Coli BL21
result: plates (LB Amp) in incubator for further processing (37 °C)
Tuesday, May 17th
SDS Gel Analysis
Investigator: CG
Aim of the experiment: SDS gel analysis of collagen 1/2, eGFP, fraction 30 of egg-precipitation
Procedure:
- mixing of 80 µl samples with 20 µl SDS buffer and heating at 95°C for 10 min. 1 d staining, 1 d unstaining
Results: successful cloning verified, stored at -20 °C
- 1. Lane: 8 µl Marker (Thermo Fisher #26610)
- 2. Lane: fraction 30 (IEC), 3 µl, band at 35 kDa, Avidin expected at 16 kDa
- 3. Lane: eGFP, 12 µl, band at 27 kDa eGFP expected at 27 kDa, many impurities
- 4. Lane: Collagen 1, 12 µl, no sharp band
- 5. Lane: Collagen 1, 12 µl, no sharp band
Minipreps pSb1C3-AviTag, -A3C5, pASK75-(SA1), -(SAm1)
Investigator: CR, CG
Aim of the experiment: Verification of cloning
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF) for pASK plasmids and 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O - 5 µl on 1% agarose gel electrophoresis of digestion
result: : successful cloning verified for pASK plasmids, repetition of pSb1C3 plasmids, stored at -20 °C
- 1. Lane: 5 µl Thermo Fisher, 1 kb Ladder
- 2. Lane: 5 µl digestion of pSb1C3-AviTag
- 3. Lane: 5 µl digestion of pSb1C3-A3C5
- 5. Lane: 5 µl digestion of pASK75(SA1), EB elution
- 6. Lane: 5 µl digestion of pASK75(SAmut1), EB elution
- 7. Lane: 5 µl digestion of pASK75(SAmut1), H2O elution
Inoculation of pre-culture with BL21 (pASK75 (SA1)) in LB-medium
Investigator: CR
Aim of the experiment: Preculture for streptavidin expression in TB-medium
Procedure:
- Add 50 µL ampicillin in 50 mL LB-medium
- Picking colonies from BL21 (pASK75 (SA1))
- Inoculate LB-medium
- Incubate at 30°C over night
Wednesday, May 18th
Repetition of analytical gel of pSb1C3-AviTag, -A3C5
Investigator: CG, CR
Aim of the experiment: Verification of cloning
Procedure:
- analytic digestion with: 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 8,5 µl Plasmid-DNA
- 10 µl on 2% agarose gel electrophoresis of digestion
Results:
Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline
Investigator: CR
Aim of the experiment: Production of streptavidin
Procedure:
- Ampicillin (2 mL) was added to the Medium (1:1000)
- The pre-culture (50 mL) was poured into the Medium
- Culture was incubated at 37°C and 140 rpm until OD550 reached 0.5
- To induce streptavidin expression anhydro-tetrazycline (200 µL) was added to the culture (1:10000)
- The culture was incubated at 37°C and 140 rpm for 4 hours
Results:
- Streptavidin expression by BL21
Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium
Investigator: CR, JB, JH
Aim of the experiment: Recombinant expression and purification of Streptavidin
Procedure:
- After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4°C, 5000 rpm, 20 mins, F 4X1L rotor)
- The supernatant was cast away and the pellet was transferred into a beaker of sufficient size and resuspended in fridge-cooled Tris Buffer B (50 mM Tris/HCl (pH = 8.0), 1 mM EDTA)
- The solution was homogenized in the PANDA (ask supervisor)
- The resulting lysate was transferred into centrifuge tubes and spun down (4°C, 18,000 rpm, 10 mins, XX34-rotor). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight.
Dialysis of eGFP
Investigator: NA, JH, CR
Aim of the experiment: Purification of eGFP
Procedure:
- eGFP was thawed on ice
- eGFP was then poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in ice cold Tris/HCl 20 mM pH 8.0
- The dialysis took place at 4°C over nightXX34-rotor). The supernatant was cast away and the pellet was
MiniPrep of quickchanged pNGAL146-A2
Investigator: NA
Aim of the experiment: Extraction of pNGAL146-A2 plasmid from XL1 blue
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
Sequencing of P14, P15 & P19
Investigator: CR, NA
Aim of the experiment: Sequencing of P14, P15 & P19
Procedure:
- Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
- The different plasmids we prepared received the following barcodes:
- P14 : FR11326653
- P15 : FR11326655
- P19 (K4): FR11326654
- P16 (K1): FR11326652
- P17 (K2): FR11326651
- P18 (K3): FR11326650
Digestion of P16, P17, P18 & P19 with AgeI & HindIII + analytical gel
Investigator: NA, JH, CR
Aim of the experiment: Verification of success of quickchange
Procedure:
- analytic digestion with: 0,25 µl HindIII (HF), 0,25 µl AgeI (HF), 1 µl SmartCut Buffer, 500 ng plasmid-DNA, fill up with ddH2O (Vtotal= 10µL)
- 10 µl on 2% agarose gel for electrophoresis
Results: No signal at 600 bp --> quickchange seems to be successful (waiting for sequencing)
Thursday, May 19th
Re-Sequencing of P19
Investigator: CR
Aim of the experiment: Re-Sequencing of P19
Procedure:Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- P19 (K4): FR11326649
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CG
Aim of the experiment: Re-Trafo of pSB1C3 RFP for later on: digestion, dephosphorylation and cloning
Procedure: transformation according to protocol of P4 E. Coli XL1
Result: plates (LB Cam) in incubator for further processing (37 °C)
Streptavidin refolding
Investigator: JB
Aim of the experiment: Refolding of denaturated Streptavidin
Procedure: After the pellet had almost completely dissolved in 6M GdmCl, the solution was spun down (4°C, 20 mins, 18,000 rpm). The supernatant was transferred carefully into a falcon tube and the pellet was cast away. Via a hydraulic pump (flow rate: 2x10 ml/min) the lysate was transferred Into 5L PBS 1x. Afterwards the pump was cleaned with technical isopropanol and ELGA water. The solution was stirred overnight at 4°C for refolding.
Biotinylation of BSA
Investigator: JB
Aim of the experiment: Biotinylation of BSA
Procedure: A 100 µM (=6.8 mg/ml) solution of BSA (Albumin fraction V, pH=7, in the fridge in the central lab) was created (V=10 ml). 220 µl of a 100 mM Biotin stock were added. The mixture was stored overnight Iin the fridge (4°C).
Result: Hopefully biotinylated BSA mixture in the fridge (4°C).
Week 2 (May 23rd - May 29th
Week 3 (May 30th - June 5th
Week 4 (June 6th - June 12th)
Week 5 (June 13th - June 19th)
Week 6 (June 20th - June 26th)
Thursday, June 23rd
Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001
Investigator: Jan, Julian
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001 Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P80 | 432,7 |
| P81 | 294,8 |
| P82 | 450,5 |
| P83 | 479,0 |
| P84 | 108,0 |
| P85 | 356,0 |
| P86 | 47,2 |
Friday, June 24th
Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2)
Investigator: Julian, Niklas, Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P82/83 (EspP K1/2) and P84/85 (StrepTag K1/2).
Procedure:
- Batch for analytical digestion for P82-P85 with EcoRI-HF
| volume | reagent |
| 0.5/1.0 µl | Plasmid DNA (-/P84) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) |
| 8/7.5 µl | ddH2O (-/P84) |
| =10 µl | TOTAL |
Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Investigator: Julian
Aim of the experiment: Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). Sequencing primer VF2 was used
The different vectors we sequenced received the following barcodes:
- mRuby3 in pSB1C3 (P80): FR11326590
- EspP in pSB1C3 (P83): FR11326588
- Streptag in pSB1C3 (P85): FR11326587
Transformation of E. coli XL1 blue with F64 (quickchanged P3(pSAm1))
Investigator: Niklas
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
--> Quickchange did not work, do again, than new transformation!
Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3)
Investigator: Niklas
Aim of the experiment: Gelextraction of F67(BirA(Digest. F59 [EcoRI; SpeI])), F68(mRuby(Digest. F60 [NgoMIV; SpeI])), F69(EGFR-TMD(Digest. F60 [NgoMIV; SpeI]), F70(pSB1C3(digest. P74 [NgoMIV; SpeI]) and F71(pSB1C3(digest. P74 [EcoRI; SpeI])
Procedure:
Gelextraction was performed by manufacturers protocol (Qiagen).
Saturday, June 25th
Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)
Investigator: Niklas
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P87 | 81 |
| P88 | 34,5 |
| P89 | 86,3 |
| P90 | 108,5 |
| P91 | 417,4 |
Analytical digestion and gelelectrophoresis of F64 (quickchanged P3) for verification of succesful quickchange
Investigator: Niklas
Procedure:
- Analytical digestion with NdeI and gelelectrophoresis. If quickchange worked there should be a band at about 3200 bp (only one restriction site left)
- Incubation over night at room temperature.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
Just a band showing a few bp (Primer), there is no plasmid band -> Quickchange did not work
Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3)and F44 + F30 (mRuby in pSB1C3)
Investigator: Niklas
Procedure:
- 6x 4 ml LB+Cam media
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
Sunday, June 26th
Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3)
Investigator: Luisa
Aim of the experiment: Extracting F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) from E.coli XL-1-blue
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P92 | 162,9 |
| P93 | 447,9 |
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- digestion of PCR-Product with DpnI for 1h at 37°C
- now labeled P94
Transformation of E. coli XL1 blue with P94 (quickchanged P3(pSAm1))
Investigator: Luisa
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) for pASK (F72) and on chloramphenicol plates for P92 and P93 and incubated over night at 37 °C in the incubator.
Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75)
Investigator: Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75).
Procedure:
- Batch for analytical digestion for P92 and P93 with EcoRI-HF and PstI-HF
| volume | reagent |
| 1 µl | Plasmid DNA |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) and PstI-HF (10 U/µl) for P92/93, NdeI (10 U/µl) for P94 |
| 7.5/7 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- 1. band (P92): no mRuby at 700bp visible, only empty vector
- 2. band (P93): empty vector and EGFR --> perfect
- 3. band (P94): no signal at all --> repetition of QC-PCR
Week 7 (June 27th - July 3rd)
Monday, June 27th
Sequencing of P67 (EGFR-Signalpeptid)
Investigator: Niklas
Procedure:
Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2))
FR11326586
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- Digestion of PCR-Product with DpnI for 1h at 37°C.
- Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR).
PCR of Genesynthesis 3 and 4
Investigator: Luisa
Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)
Procedure:
- The PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 2,5 µl | Primer VF2 |
| 2,5 µl | Primer VR2 |
| 1 µl | dNTP-mix |
| 10 µl | Q5 Polymerase reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Q5-Polymerase |
| 18 µl | ddH2O |
- Setup: iGEM_standard (Promega-cycler)
| temperature | time |
| 98°C | 2min |
| 98°C | 10sec |
| 66°C | 30sec |
| 72°C | 30sec |
| 72°C | 2min |
| 4°C | hold |
- the batches were then purified using the Quiagen PCR-Purification Kit.
Analytical digestion and gelelectrophoresis of P88 , P89 and P90
Investigator: Niklas
Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)
Procedure:
- Batches for analytical digestions:
P88: EcoRI
P89: EcoRI and PstI
P90: EcoRI
| volume | reagent |
| 5,8/2,3/1,8 µl | Plasmid DNA (P88/P89/P90) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl)/ PstI |
| required amount for total volume of 10 µl | ddH2O |
Ligation of F67 and F71, Transformation of E. coli XL1 blue afterwards
Investigator: Niklas
Aim of the experiment: Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 2,4 µl | Vektor |
| 7,6 µl | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
- next step: analytic digestion of transformation was successful
Digestion of PCR on genesynthesis 3 and 4, and pSB1C3
Investigator: Luisa
Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.
Procedure:
- Batches for analytical digestions:
| volume | reagent |
| 1 µl each | enzyme (SapI, HindIII, XbaI, AgeI) |
| 5 µl | CutSmart buffer (10x) |
| 41 µl | DNA (purified PCR-products of GSY3 and 4) |
- Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80.
Analytical digestion and gelelectrophoresis of P80 , P78 and P85
Investigator: Julian
Aim of experiment: Analytical digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85(Strep-Tag)
Procedure:
- Batches for analytical digestions:
- P80 and P78: EcoRI and AgeI
- P85: EcoRI and NgoMIV
| volume | reagent |
| 10 µl | Plasmid DNA |
| 32 µl | ddH2O |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme(10 U/µl)/ PstI |
| 50 µl | TOTAL |
Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into E. coli XL1 blue
Investigator: Luisa
Aim of the experiment: Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 4,3µl(for F75), 8,2µl (for F76) | Vector |
| 12,7µl (F75), 8,8 (F76) | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| =20 µl | TOTAL |
- Ligation was incubated at RT for 1,5h.
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator.
Chemical biotinylation of BSA
Investigator: Niklas
Procedure:
- BSA was chemically biotinylated with a 20x and 40x molar excess:
- 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85)
- dissolve BSA (10 mg/ml)
- Add biotin-NHS-ester: 20,5 mg for 40x molar excess
- reaction over night