Gracetexana (Talk | contribs) |
Gracetexana (Talk | contribs) |
||
| Line 142: | Line 142: | ||
The first steps in the characterization of microbes native to kombucha involved the isolation of strains from store-bought kombucha samples. This was accomplished by plating various dilutions of kombucha onto a variety of media including YPD, HS, and R2A. | The first steps in the characterization of microbes native to kombucha involved the isolation of strains from store-bought kombucha samples. This was accomplished by plating various dilutions of kombucha onto a variety of media including YPD, HS, and R2A. | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/0/00/T--Austin_UTexas--IsolatingStrains.png" alt="RecapitulationsDay1" style="width: | + | <center><img src="https://static.igem.org/mediawiki/2016/0/00/T--Austin_UTexas--IsolatingStrains.png" alt="RecapitulationsDay1" style="width:80%;"></center> |
<figcaption><b>Figure 1:</b>Shows YPD plates spread with various dilutions of GT's brand kombucha samples. | <figcaption><b>Figure 1:</b>Shows YPD plates spread with various dilutions of GT's brand kombucha samples. | ||
</figure> | </figure> | ||
| Line 150: | Line 150: | ||
</p> | </p> | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/4/44/T--Austin_UTexas--ExampleGel2.png" alt="RecapitulationsDay1" style="width: | + | <center><img src="https://static.igem.org/mediawiki/2016/4/44/T--Austin_UTexas--ExampleGel2.png" alt="RecapitulationsDay1" style="width:80%;"></center> |
<figcaption><b>Figure 2:</b>Example of a gel obtained after running PCR products. Lane #1 is a 100 bp ladder; Lanes 2-7 are six different kombucha isolates that underwent PCR reactions selecting for bacteria (targeting 16S rRNA gene); Lanes 8-13 are the same six isolates, in the same order, but which underwent reactions selecting for fungi (targeting ITS rRNA gene). Based on the gel results it can be observed that for any given isolate, a product in gel was only observed for either bacterial or fungal primer reactions. | <figcaption><b>Figure 2:</b>Example of a gel obtained after running PCR products. Lane #1 is a 100 bp ladder; Lanes 2-7 are six different kombucha isolates that underwent PCR reactions selecting for bacteria (targeting 16S rRNA gene); Lanes 8-13 are the same six isolates, in the same order, but which underwent reactions selecting for fungi (targeting ITS rRNA gene). Based on the gel results it can be observed that for any given isolate, a product in gel was only observed for either bacterial or fungal primer reactions. | ||
</figure> | </figure> | ||
| Line 157: | Line 157: | ||
Once it was determined whether each isolate was a bacterium or a fungus, the PCR products were purified and samples of the gDNA was sequenced using Sanger sequencing. The resulting sequences were then ran through the Ribosomal Database Project (RDP) SeqMatch tool in order to identify the exact species of bacteria or yeast that correspond to each tested isolate. The identified microbes are listed below in Table 1. | Once it was determined whether each isolate was a bacterium or a fungus, the PCR products were purified and samples of the gDNA was sequenced using Sanger sequencing. The resulting sequences were then ran through the Ribosomal Database Project (RDP) SeqMatch tool in order to identify the exact species of bacteria or yeast that correspond to each tested isolate. The identified microbes are listed below in Table 1. | ||
</p> | </p> | ||
| + | <center> | ||
</html> | </html> | ||
| − | {| class="wikitable" style="width: | + | {| class="wikitable" style="width: 80%;" |
|+Table 1:Microbes Isolated and Identified from Various Store Bought Kombucha Samples | |+Table 1:Microbes Isolated and Identified from Various Store Bought Kombucha Samples | ||
! |Species | ! |Species | ||
| Line 203: | Line 204: | ||
(*Indicates a species that is considered vital to the production of kombucha) | (*Indicates a species that is considered vital to the production of kombucha) | ||
<html> | <html> | ||
| + | </center> | ||
</div> | </div> | ||
| Line 210: | Line 212: | ||
<h2>Recapitulation</h2> | <h2>Recapitulation</h2> | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/3/38/T--Austin_UTexas--PreviousRecaps.png" alt="RecapitulationsDay1" style="width: | + | <center><img src="https://static.igem.org/mediawiki/2016/3/38/T--Austin_UTexas--PreviousRecaps.png" alt="RecapitulationsDay1" style="width:80%;"></center> |
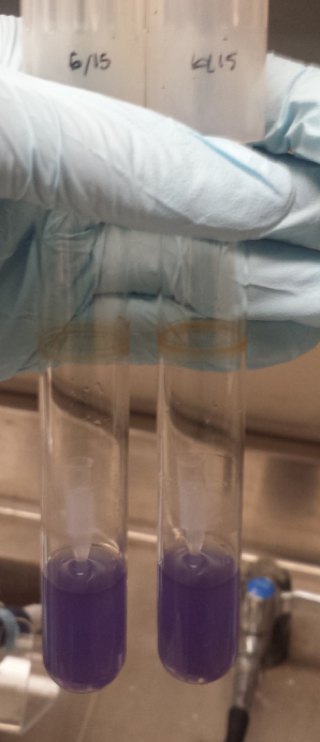
<figcaption><b>Figure 1:</b>Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls. | <figcaption><b>Figure 1:</b>Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls. | ||
</figure> | </figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/d/d6/T--Austin_UTexas--OngoingRecapitulations.png" alt="Conjugation2" style="width: | + | <center><img src="https://static.igem.org/mediawiki/2016/d/d6/T--Austin_UTexas--OngoingRecapitulations.png" alt="Conjugation2" style="width:80%;"></center> |
</div> | </div> | ||
| Line 255: | Line 257: | ||
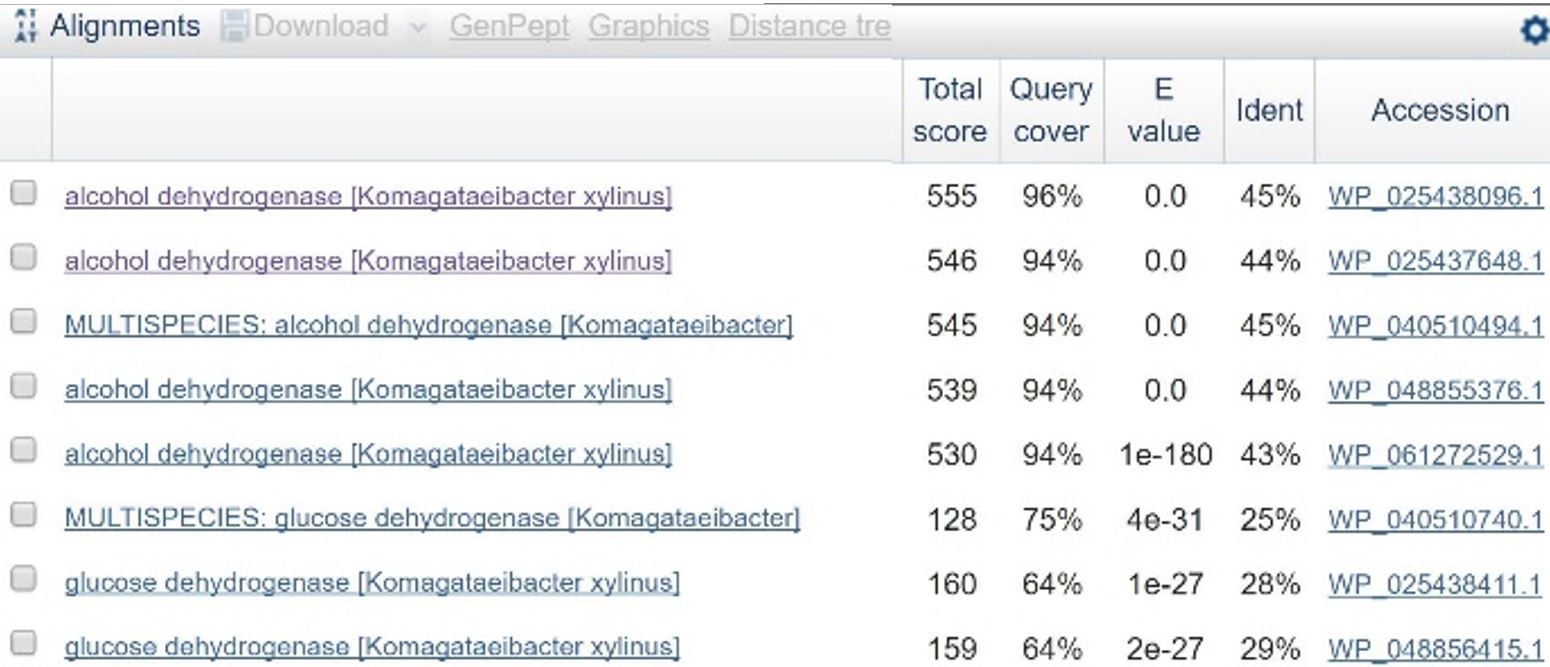
<h4>GOX Sequences as Putative Promoters</h4> | <h4>GOX Sequences as Putative Promoters</h4> | ||
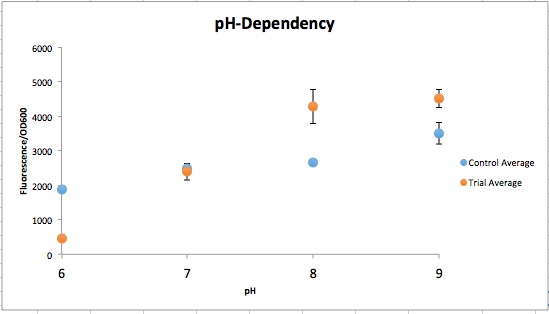
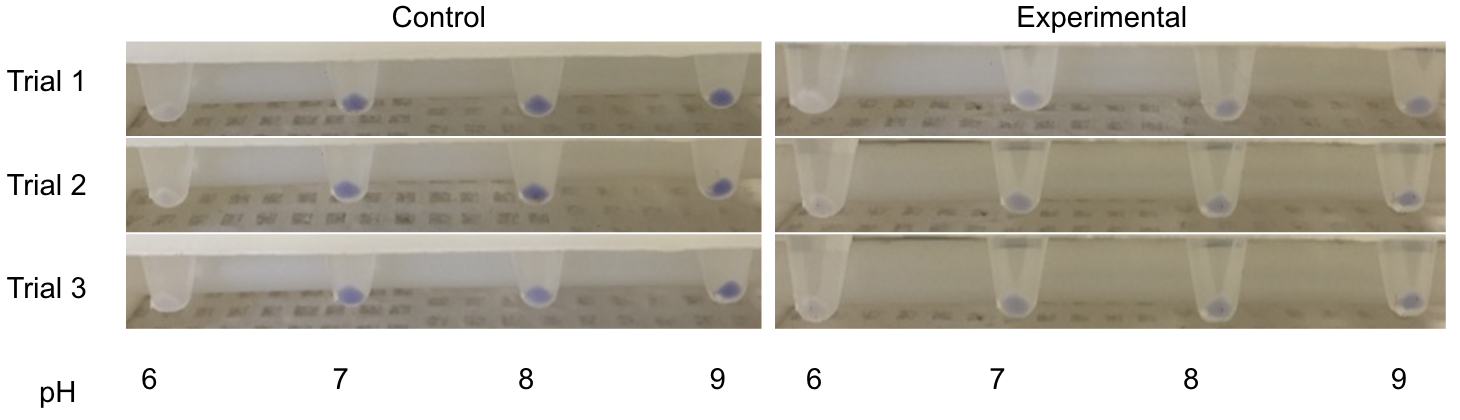
<p>Three endogenous upstream regions of loci that were reported to show increased mRNA synthesis as pH decreased were obtained. Using Golden Gate assembly, these putative promoters will be placed on a plasmid with a specific reporter sequence.<sup>2</sup> By placing these pH-sensitive promoters with different reporters and transforming into multiple organisms, the visualization of the microbes and their location in kombucha would be possible. This would serve as a stepping stone into the transformation of multiple kombucha organisms with these different reporter constructs, meaning organism concentration at a specific time during the brewing process could be visualized.</p> | <p>Three endogenous upstream regions of loci that were reported to show increased mRNA synthesis as pH decreased were obtained. Using Golden Gate assembly, these putative promoters will be placed on a plasmid with a specific reporter sequence.<sup>2</sup> By placing these pH-sensitive promoters with different reporters and transforming into multiple organisms, the visualization of the microbes and their location in kombucha would be possible. This would serve as a stepping stone into the transformation of multiple kombucha organisms with these different reporter constructs, meaning organism concentration at a specific time during the brewing process could be visualized.</p> | ||
| + | <center> | ||
</html> | </html> | ||
| − | {| class="wikitable" style="width: | + | {| class="wikitable" style="width: 80%;" |
|+Table 1:The Three Endogenous GOX Sequences | |+Table 1:The Three Endogenous GOX Sequences | ||
! |Locus Tag | ! |Locus Tag | ||
| Line 275: | Line 278: | ||
|3.36 | |3.36 | ||
|} | |} | ||
| − | + | <html> | |
| − | + | </center> | |
<br><br><br><br><br> | <br><br><br><br><br> | ||
<h3>References</h3> | <h3>References</h3> | ||
<ol type="1"> | <ol type="1"> | ||
| − | + | <li><a href="https://2015.igem.org/Team:BIT-China/Parts">BIT-China-2015</a></li></html> | |
<li>Hanke, T., Richhardt, J., Polen, T., Sahm, H., Bringer, S., and Bott, M. (2012) Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. <i>Journal of Biotechnology 157</i>, 359–372.</li> | <li>Hanke, T., Richhardt, J., Polen, T., Sahm, H., Bringer, S., and Bott, M. (2012) Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. <i>Journal of Biotechnology 157</i>, 359–372.</li> | ||
<li>Kuper, C., and Jung, K. (2005) CadC-mediated activation of the cadBA promoter in Escherichia coli. <i>Journal of Molecular and Microbiological Biotechnology 1</i>, 26–39.</li> | <li>Kuper, C., and Jung, K. (2005) CadC-mediated activation of the cadBA promoter in Escherichia coli. <i>Journal of Molecular and Microbiological Biotechnology 1</i>, 26–39.</li> | ||
Revision as of 17:28, 18 October 2016