Ianoverman (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
div.naviSection { | div.naviSection { | ||
| − | |||
} | } | ||
| Line 17: | Line 16: | ||
</head> | </head> | ||
<body> | <body> | ||
| − | <div class = "column | + | |
| + | <div class = "column full_size_outer"> | ||
| + | <div class = "column_full_size_inner"> | ||
<h1>Austin_UTexas<h1> | <h1>Austin_UTexas<h1> | ||
<h2>Results</h2> | <h2>Results</h2> | ||
| Line 45: | Line 46: | ||
<input type="image" src ="https://static.igem.org/mediawiki/2016/e/e7/T--Austin_UTexas--pHNavi.png" style="width:100%;" onclick="showOne('section6')" /><p>pH Sensors</p> | <input type="image" src ="https://static.igem.org/mediawiki/2016/e/e7/T--Austin_UTexas--pHNavi.png" style="width:100%;" onclick="showOne('section6')" /><p>pH Sensors</p> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 206: | Line 119: | ||
</div> | </div> | ||
| + | <div class="naviSection" id="section2"> | ||
| + | <h2>Conjugation</h2> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2016/3/3f/T--Austin_UTexas--MIC.png" alt="RecapitulationsDay1" style="width:80%;"></center> | ||
| + | <p>We have attempted to conjugate GFP into both <i>G. oxydans</i> and <i>G. hansenii</i> with a Diaminopimelic Acid (DAP) auxotrophic strain of <i> E. coli </i>. The plasmid contains the vector pMMB67EH, the promoter PA-1, GFP and a spectinomycin resistance gene. </p> | ||
| + | <p>The first conjugation was done with KOM strains 4 (<i>G. oxydans</i>) 5 (<i> G. oxydans </i> and 15 (<i> L. fermentati </i>). We attempted these conjugations before sequencing the recipient strains, so that is why we tried to conjugate into <i> L. fermentati </i>. First, a mixture between a KOM strain and the DAP auxotroph strain were plated on a LB+DAP solid medium to allow for conjugation to occur. After 24 hours of incubation, we scraped up the growth and plated each conjugation mixture onto a LB+Spec plate.</p> | ||
| + | </html> | ||
| + | |||
| + | [[File:T--Austin UTexas--LB+SPEC1stconjugation.jpg|thumb|left|300px|LB+SPEC plates that contain conjugation mixtures of KOM 4, 5 and 15 (<i> L. fermentati </i>)]] | ||
| + | |||
| + | <html> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <p>Next, we viewed the potential transconjugants on a fluorescence microscope.</p> | ||
| + | </html> | ||
| + | [[File:T--Austin UTexas--KOM4nogfp.png|thumb|left|600px|Using a fluorescent microscope, this was a picture taken of the <i>G. oxydans</i> strain, KOM 4, without the plasmid that contains GFP.]] | ||
| + | |||
| + | [[File:T--Austin UTexas--KOM4withgfpandcirclesfixed.png|thumb|left|600px|Using a fluorescent microscope, this was a picture taken of the potential transconjugant, KOM 4, with the plasmid GFP. '''16s sequencing was still needed to confirm successful conjugation.''']] | ||
| + | <html> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br> | ||
| + | <p>We then picked these glowing colonies and then after streaking them out onto more LB+Spec plates, we attempted to use 16s sequencing to confirm successful conjugation. After troubleshooting our 16s procedure, we were finally able to obtain a viable sequencing result. However, all of the glowing colonies were identified as <i>E. coli</i>. For the next round of conjugation, we used a strain of both <i>G. oxydans</i> and <i>Gluconacetobacter hansenii</i> from the American Type Culture Collection (ATCC). | ||
| + | </html> | ||
| + | [[File:T--Austin UTexas--FixedLB+DAP2ndconj.jpeg|thumb|left|300px|This is a LB+DAP plate on a dark reader that has four different conjugations occurring at one time. The two left quadrants have the same ATCC strain of <i>G. oxydans,</i>, while the two quadrants on the right have <i>Ga. hansenii</i>.]] | ||
| + | <html> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br> | ||
| + | <p>These growths were then scraped up and plated onto a LB+Spec plate.</p> | ||
| + | </html> | ||
| + | [[File:T--Austin UTexas--LB+SPEC2ndconj.jpg|thumb|left|300px|These are my potential transconjugants on a LB+DAP plates. The dark reader was used when taking this picture. The top two are <i>G. oxydans</i> while the bottom two are <i>G. hansenii</i>.]] | ||
| + | <html> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <p>We then picked isolated colonies and streaked them out onto LB+DAP plates. After using 16s sequencing on the potential transconjugants, we encountered an anomaly. Instead of amplifying the 16s gene, we recieved the sequence of the L,D Transpeptidase gene of <i> E. coli </i>. We plan on repeating the 16s procedure.</p> | ||
| + | <p> | ||
| + | </div> | ||
<div class="naviSection" id="section3"> | <div class="naviSection" id="section3"> | ||
| Line 222: | Line 196: | ||
</figure> | </figure> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | <div class="naviSection" id="section4"> | ||
| + | <h2>Ethanol Reduction</h2> | ||
| + | </html> | ||
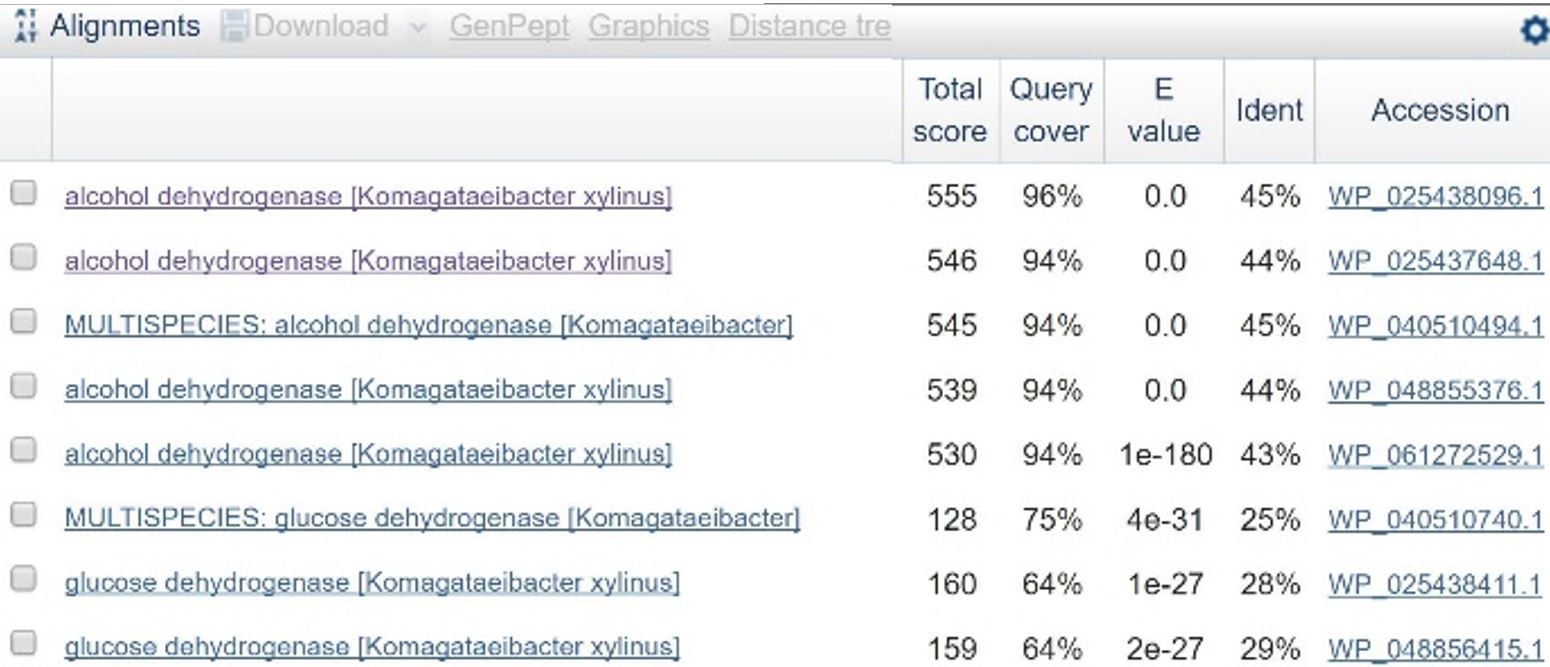
| + | [[File:T-Austin UTexas--File ADHblasttable.png|thumb|left|'''Table 1''': Results of BLAST search comparing the amino acid sequence for PQQ-ADH in ''C. testosteroni'' against similar amino acid sequences in ''Komagataeibacter xylinus'' (identical to ''Ga. hansenii''). Line 3 is a close match, and the accession number matches one of the ADH genes found in ''K. xylinus''. |600px]] | ||
| + | [[File:T--Austin UTexas--ALDHlinmap.png|thumb|left|'''Figure 1''': Linear map of the coding sequence for membrane-bound ALDH with a Golden Gate type 3 prefix and suffix. BsmBI and BsaI sites are indicated. The restriction sites at either end are included in the prefix and suffix, but the internal BsaI site must be removed to create a functioning Golden Gate part.|600px]] | ||
| + | [[File:T--Austin UTexas--ADHlinmap.png|thumb|left|'''Figure 2''': Linear map of the coding sequence for PQQ-ADH with a Golden Gate type 3 prefix and suffix. EcoRI, BsmBI, and BsaI sites are indicated. The centermost BsmBI restriction site is in the coding sequence and must be removed to create a functional Golden Gate part. EcoRI is not used in Golden Gate assembly, so those sites do not necessarily need to be removed.|600px]] | ||
| + | <html> | ||
| + | <p> | ||
| + | <u>Identifying genes of interest</u> | ||
| + | <p> | ||
| + | In order to design a construct increasing expression of PQQ-ADH and ALDH in <i>Ga. hansenii</i>, it was necessary to find the genome of the ATCC strain and identify the coding sequences for these genes. The whole genome shotgun sequence for our organism, ATCC 53582, is published on NCBI by J. Abbot (2015) with annotations regarding the functions of specific sequences. Coding sequences are annotated with proposed gene products. Though there are several aldehyde dehydrogenase genes annotated in the genome, there is only one which is described as membrane-bound, matching the description from Mamlouk and Gullo (2013). There are additionally multiple alcohol dehydrogenases. A known amino acid sequence for a homologous PQQ-ADH in ,i.Comamonas testosteroni</i> was compared against sequences in the <i>Ga. hansenii</i> genome using BLAST (<b>Table 1</b>). One ADH enzyme found in the <i>Ga. hansenii</i> genome sequence matches the <i>C. testosteroni</i> sequence with a query cover value of 94% and an E value of 0 (third line of <b>Table 1</b>).</p> | ||
| + | <p><u>Creation of Golden Gate parts</u> | ||
| + | <p> | ||
| + | In order to assemble the construct, the coding sequences for the genes of interest must be amplified from the <i>Ga. hansenii</i> genome and edited such that they have the correct Golden Gate overhangs and no internal BsaI or BsmBI restriction sites. The sequences were uploaded to Benchling for analysis and planning. The coding sequence for the membrane-bound ALDH contains a BsaI restriction site near the middle of the gene (<b>Figure 1</b>), and the PQQ-ADH coding sequence contains a BsmBI restriction site near the end of the gene (<b>Figure 2</b>). To eliminate the BsaI site in ALDH, primers were designed that would introduce a point mutation at the restriction site. One set of primers, igem2016_KOM_EtOH_01 and igem2016_KOM_EtOH_02, amplifies the sequence upstream of the restriction site, adding a type 3 Golden Gate prefix and removing the restriction site. Another set, igem2016_KOM_EtOH_03 and igem2016_KOM_EtOH_04, amplifies the region downstream of the restriction site, introducing a mutation to the site and adding a type 3 Golden Gate suffix to the end of the gene. These two products will be used in an overlap PCR reaction to create a final product with no BsaI restriction sites and the correct prefix and suffix for assembly. To remove the BsmBI site from the PQQ-ADH coding sequence, a set of primers (igem2016_KOM_EtOH_05 and igem2016_KOM_EtOH_06) was designed to amplify the region upstream of the restriction site and add a Golden Gate type 3 prefix to the beginning of the sequence. The reverse primer additionally adds a mutation to existing BsmBI restriction site and creates a new BsmBI restriction site that will be used to join the piece to a double-stranded DNA, igem2016_KOM_EtOH_07, containing the rest of the gene’s coding sequence appended with a Golden Gate type 3 suffix. The assembly of the PQQ-ADH part will therefore take place in two reactions: one reaction in which the upstream piece of DNA is created, and one reaction in which it is ligated to the gBlock. <b>Table 2</b> contains more information about each of these oligonucleotides. All were ordered from IDT. | ||
| + | </html> | ||
| + | [[File:T--Austin UTexas--oligotable.png|thumb|center|'''Table 2''': Description of oligonucleotides ordered from IDT and their purposes. All of these are PCR primers except for igem2016_KOM_EtOH_07, which is a gBlock containing the end of the PQQ-ADH with a Golden Gate type 3 suffix appended.|600px]] | ||
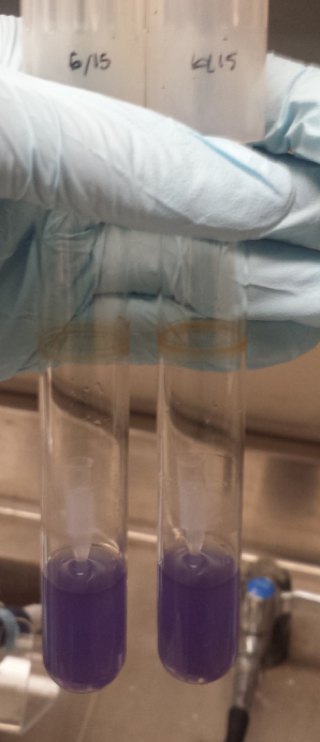
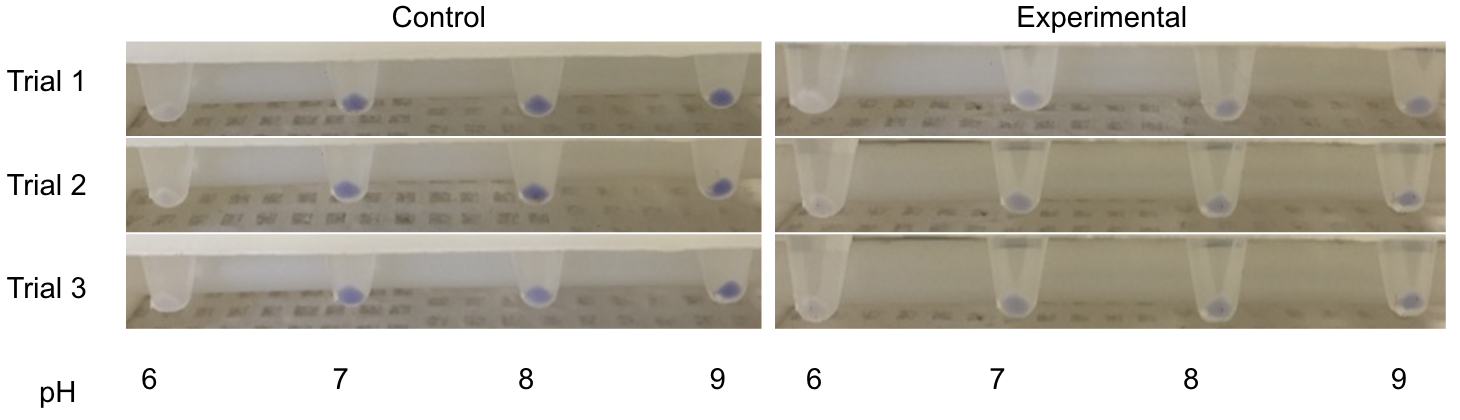
| + | [[File:T--Austin UTexas--bromothymolblue wkey.png|thumb|'''Figure 3''': YPD plates made with pH indicator bromothymol blue. Colonies are various strains of ''Lachancea fermentati'' isolated from kombucha in our lab. Carbon dioxide and ethanol form as products of fermentation. The carbon dioxide reacts with water to form carbonic acid, lowering the pH of the plate and changing the color of the pH indicator. More dramatic color changes should correlate to greater ethanol production, but this assay is limited in that a variety of metabolites unrelated to ethanol production could influence pH.|400px]]<html> | ||
| + | </div> | ||
| + | |||
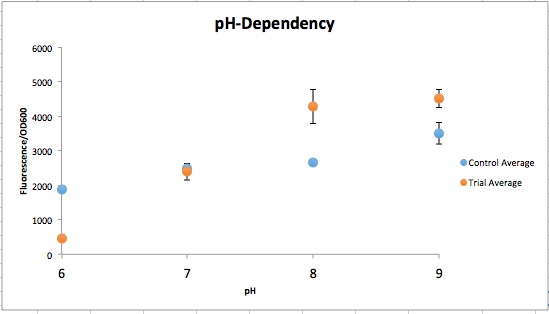
<div class="naviSection" id="section6"> | <div class="naviSection" id="section6"> | ||
| Line 287: | Line 282: | ||
<li>Nakayama, S.-I., and Watanabe, H. (1998) Identification of cpxR as a Positive Regulator Essential for Expression of the Shigella sonnei virF Gene. <i>Journal of Bacteriology 180</i>, 3522–3528.</li> | <li>Nakayama, S.-I., and Watanabe, H. (1998) Identification of cpxR as a Positive Regulator Essential for Expression of the Shigella sonnei virF Gene. <i>Journal of Bacteriology 180</i>, 3522–3528.</li> | ||
<li>Nakayama, S.-I., and Watanabe, H. (1995) Involvement of cpxA, a Sensor of a Two-Component Regulatory System, in the pH-Dependent Regulation of Expression of Shigella sonnei virF Gene. <i>Journal of Bacteriology 177</i>, 5062–5069.</li> | <li>Nakayama, S.-I., and Watanabe, H. (1995) Involvement of cpxA, a Sensor of a Two-Component Regulatory System, in the pH-Dependent Regulation of Expression of Shigella sonnei virF Gene. <i>Journal of Bacteriology 177</i>, 5062–5069.</li> | ||
| + | </div> | ||
| + | </div> | ||
Revision as of 23:17, 18 October 2016
Austin_UTexas
Results

Results

Click on one of the images below to learn more about our results!
Kombucha Strains
Conjugation
Recapitulation
Ethanol
pH Sensors