Labjournal
Samples
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with
Investigator:
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
Week 1, May 16th - May 22nd
Monday, May 16th
Streptavidin plasmids control
Investigator: JB, LK, JH
Aim of the experiment: Verification of cloning
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) (4 clones each of pSA1, pSAm1 in pASK75)
- analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF), 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O
- 5 µl on 1% agarose gel electrophoresis of digestion
Results: successful cloning verified, stored at -20 °C
- 1. Lane: 5 µl Thermo Fisher, 1kb Ladder
- 2. to 9. Lane: 5 µl digestions of P6 to P13, band of SA (mut1) at about 300 bp, band of digested plasmid at about 3.000 bp
Streptavidin expression_trafo BL21
Investigator: JB, JH
Aim of the experiment: expression of pSA1 and pSAm1 in E. Coli BL21
Procedure:
- transformation according to protocol of P6 and P10 in competent E. Coli BL21
result: plates (LB Amp) in incubator for further processing (37 °C)
Tuesday, May 17th
SDS Gel Analysis
Investigator: CG
Aim of the experiment: SDS gel analysis of collagen 1/2, eGFP, fraction 30 of egg-precipitation
Procedure:
- mixing of 80 µl samples with 20 µl SDS buffer and heating at 95°C for 10 min. 1 d staining, 1 d unstaining
Results: successful cloning verified, stored at -20 °C
- 1. Lane: 8 µl Marker (Thermo Fisher #26610)
- 2. Lane: fraction 30 (IEC), 3 µl, band at 35 kDa, Avidin expected at 16 kDa
- 3. Lane: eGFP, 12 µl, band at 27 kDa eGFP expected at 27 kDa, many impurities
- 4. Lane: Collagen 1, 12 µl, no sharp band
- 5. Lane: Collagen 1, 12 µl, no sharp band
Minipreps pSb1C3-AviTag, -A3C5, pASK75-(SA1), -(SAm1)
Investigator: CR, CG
Aim of the experiment: Verification of cloning
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- analytic digestion with: 0,25 µl XbaI, 0,25 µl HindIII (HF) for pASK plasmids and 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 5 µl Plasmid-DNA, 3,5 µl H2O - 5 µl on 1% agarose gel electrophoresis of digestion
result: : successful cloning verified for pASK plasmids, repetition of pSb1C3 plasmids, stored at -20 °C
- 1. Lane: 5 µl Thermo Fisher, 1 kb Ladder
- 2. Lane: 5 µl digestion of pSb1C3-AviTag
- 3. Lane: 5 µl digestion of pSb1C3-A3C5
- 5. Lane: 5 µl digestion of pASK75(SA1), EB elution
- 6. Lane: 5 µl digestion of pASK75(SAmut1), EB elution
- 7. Lane: 5 µl digestion of pASK75(SAmut1), H2O elution
Inoculation of pre-culture with BL21 (pASK75 (SA1)) in LB-medium
Investigator: CR
Aim of the experiment: Preculture for streptavidin expression in TB-medium
Procedure:
- Add 50 µL ampicillin in 50 mL LB-medium
- Picking colonies from BL21 (pASK75 (SA1))
- Inoculate LB-medium
- Incubate at 30°C over night
Wednesday, May 18th
Repetition of analytical gel of pSb1C3-AviTag, -A3C5
Investigator: CG, CR
Aim of the experiment: Verification of cloning
Procedure:
- analytic digestion with: 0,25 µl NgomIV, 0,25 µl AgeI (HF) for pSb1C3 plasmids, 1 µl SmartCut Buffer, 8,5 µl Plasmid-DNA
- 10 µl on 2% agarose gel electrophoresis of digestion
Results:
Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline
Investigator: CR
Aim of the experiment: Production of streptavidin
Procedure:
- Ampicillin (2 mL) was added to the Medium (1:1000)
- The pre-culture (50 mL) was poured into the Medium
- Culture was incubated at 37°C and 140 rpm until OD550 reached 0.5
- To induce streptavidin expression anhydro-tetrazycline (200 µL) was added to the culture (1:10000)
- The culture was incubated at 37°C and 140 rpm for 4 hours
Results:
- Streptavidin expression by BL21
Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium
Investigator: CR, JB, JH
Aim of the experiment: Recombinant expression and purification of Streptavidin
Procedure:
- After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4°C, 5000 rpm, 20 mins, F 4X1L rotor)
- The supernatant was cast away and the pellet was transferred into a beaker of sufficient size and resuspended in fridge-cooled Tris Buffer B (50 mM Tris/HCl (pH = 8.0), 1 mM EDTA)
- The solution was homogenized in the PANDA (ask supervisor)
- The resulting lysate was transferred into centrifuge tubes and spun down (4°C, 18,000 rpm, 10 mins, XX34-rotor). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight.
Dialysis of eGFP
Investigator: NA, JH, CR
Aim of the experiment: Purification of eGFP
Procedure:
- eGFP was thawed on ice
- eGFP was then poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in ice cold Tris/HCl 20 mM pH 8.0
- The dialysis took place at 4°C over nightXX34-rotor). The supernatant was cast away and the pellet was
MiniPrep of quickchanged pNGAL146-A2
Investigator: NA
Aim of the experiment: Extraction of pNGAL146-A2 plasmid from XL1 blue
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
Sequencing of P14, P15 & P19
Investigator: CR, NA
Aim of the experiment: Sequencing of P14, P15 & P19
Procedure:
- Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
- The different plasmids we prepared received the following barcodes:
- P14 : FR11326653
- P15 : FR11326655
- P19 (K4): FR11326654
- P16 (K1): FR11326652
- P17 (K2): FR11326651
- P18 (K3): FR11326650
Digestion of P16, P17, P18 & P19 with AgeI & HindIII + analytical gel
Investigator: NA, JH, CR
Aim of the experiment: Verification of success of quickchange
Procedure:
- analytic digestion with: 0,25 µl HindIII (HF), 0,25 µl AgeI (HF), 1 µl SmartCut Buffer, 500 ng plasmid-DNA, fill up with ddH2O (Vtotal= 10µL)
- 10 µl on 2% agarose gel for electrophoresis
Results: No signal at 600 bp --> quickchange seems to be successful (waiting for sequencing)
Thursday, May 19th
Re-Sequencing of P19
Investigator: CR
Aim of the experiment: Re-Sequencing of P19
Procedure:Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- P19 (K4): FR11326649
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CG
Aim of the experiment: Re-Trafo of pSB1C3 RFP for later on: digestion, dephosphorylation and cloning
Procedure: transformation according to protocol of P4 E. Coli XL1
Result: plates (LB Cam) in incubator for further processing (37 °C)
Streptavidin refolding
Investigator: JB
Aim of the experiment: Refolding of denaturated Streptavidin
Procedure: After the pellet had almost completely dissolved in 6M GdmCl, the solution was spun down (4°C, 20 mins, 18,000 rpm). The supernatant was transferred carefully into a falcon tube and the pellet was cast away. Via a hydraulic pump (flow rate: 2x10 ml/min) the lysate was transferred Into 5L PBS 1x. Afterwards the pump was cleaned with technical isopropanol and ELGA water. The solution was stirred overnight at 4°C for refolding.
Biotinylation of BSA
Investigator: JB
Aim of the experiment: Biotinylation of BSA
Procedure: A 100 µM (=6.8 mg/ml) solution of BSA (Albumin fraction V, pH=7, in the fridge in the central lab) was created (V=10 ml). 220 µl of a 100 mM Biotin stock were added. The mixture was stored overnight Iin the fridge (4°C).
Result: Hopefully biotinylated BSA mixture in the fridge (4°C).
Week 2 (May 23rd - May 29th)
Week 3 (May 30th - June 5th)
Week 4 (June 6th - June 12th)
Week 5 (June 13th - June 19th)
Week 6 (June 20th - June 26th)
Thursday, June 23rd
Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001
Investigator: Jan, Julian
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001 Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P80 | 432,7 |
| P81 | 294,8 |
| P82 | 450,5 |
| P83 | 479,0 |
| P84 | 108,0 |
| P85 | 356,0 |
| P86 | 47,2 |
Friday, June 24th
Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2)
Investigator: Julian, Niklas, Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P82/83 (EspP K1/2) and P84/85 (StrepTag K1/2).
Procedure:
- Batch for analytical digestion for P82-P85 with EcoRI-HF
| volume | reagent |
| 0.5/1.0 µl | Plasmid DNA (-/P84) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) |
| 8/7.5 µl | ddH2O (-/P84) |
| =10 µl | TOTAL |
Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Investigator: Julian
Aim of the experiment: Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). Sequencing primer VF2 was used
The different vectors we sequenced received the following barcodes:
- mRuby3 in pSB1C3 (P80): FR11326590
- EspP in pSB1C3 (P83): FR11326588
- Streptag in pSB1C3 (P85): FR11326587
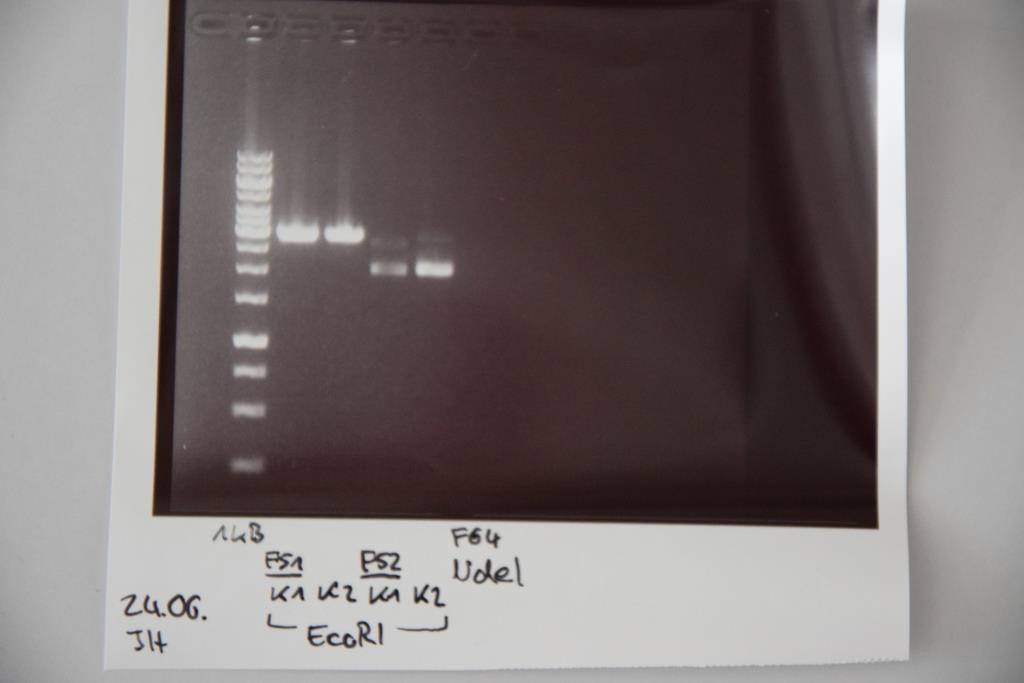
Transformation of E. coli XL1 blue with F64 (quickchanged P3(pSAm1))
Investigator: Niklas
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C.
Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3)
Investigator: Niklas
Aim of the experiment: Gelextraction of F67(BirA(Digest. F59 [EcoRI; SpeI])), F68(mRuby(Digest. F60 [NgoMIV; SpeI])), F69(EGFR-TMD(Digest. F60 [NgoMIV; SpeI]), F70(pSB1C3(digest. P74 [NgoMIV; SpeI]) and F71(pSB1C3(digest. P74 [EcoRI; SpeI])
Procedure:
Gelextraction was performed by manufacturers protocol (Qiagen).
Saturday, June 25th
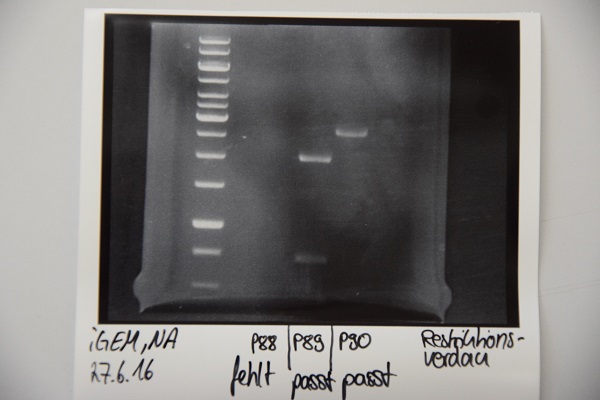
Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)
Investigator: Niklas
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P87 | 81 |
| P88 | 34,5 |
| P89 | 86,3 |
| P90 | 108,5 |
| P91 | 417,4 |
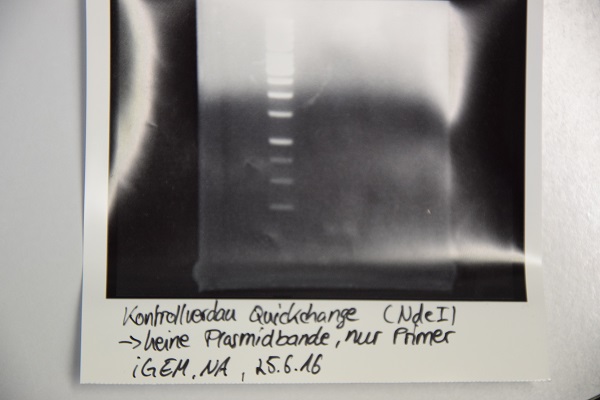
Analytical digestion and gelelectrophoresis of F64 (quickchanged P3) for verification of succesful quickchange
Investigator: Niklas
Procedure:
- Analytical digestion with NdeI and gelelectrophoresis. If quickchange worked there should be a band at about 3200 bp (only one restriction site left)
- Incubation over night at room temperature.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
Just a band showing a few bp (Primer), there is no plasmid band -> Quickchange did not work
Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3)and F44 + F30 (mRuby in pSB1C3)
Investigator: Niklas
Procedure:
- 6x 4 ml LB+Cam media
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
Sunday, June 26th
Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3)
Investigator: Luisa
Aim of the experiment: Extracting F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) from E.coli XL-1-blue
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P92 | 162,9 |
| P93 | 447,9 |
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- digestion of PCR-Product with DpnI for 1h at 37°C
- now labeled P94
Transformation of E. coli XL1 blue with P94 (quickchanged P3(pSAm1))
Investigator: Luisa
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) for pASK (F72) and on chloramphenicol plates for P92 and P93 and incubated over night at 37 °C in the incubator.
Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75)
Investigator: Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75).
Procedure:
- Batch for analytical digestion for P92 and P93 with EcoRI-HF and PstI-HF
| volume | reagent |
| 1 µl | Plasmid DNA |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) and PstI-HF (10 U/µl) for P92/93, NdeI (10 U/µl) for P94 |
| 7.5/7 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- 1. band (P92): no mRuby at 700bp visible, only empty vector
- 2. band (P93): empty vector and EGFR --> perfect
- 3. band (P94): no signal at all --> repetition of QC-PCR
Week 7 (June 27th - July 3rd)
Monday, June 27th
Sequencing of P67 (EGFR-Signalpeptid)
Investigator: Niklas
Procedure:
Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2))
FR11326586
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- Digestion of PCR-Product with DpnI for 1h at 37°C.
- Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR).
PCR of Genesynthesis 3 and 4
Investigator: Luisa
Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)
Procedure:
- The PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 2,5 µl | Primer VF2 |
| 2,5 µl | Primer VR2 |
| 1 µl | dNTP-mix |
| 10 µl | Q5 Polymerase reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Q5-Polymerase |
| 18 µl | ddH2O |
- Setup: iGEM_standard (Promega-cycler)
| temperature | time |
| 98°C | 2min |
| 98°C | 10sec |
| 66°C | 30sec |
| 72°C | 30sec |
| 72°C | 2min |
| 4°C | hold |
- the batches were then purified using the Quiagen PCR-Purification Kit.
Analytical digestion and gelelectrophoresis of P88 , P89 and P90
Investigator: Niklas
Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)
Procedure:
- Batches for analytical digestions:
P88: EcoRI
P89: EcoRI and PstI
P90: EcoRI
| volume | reagent |
| 5,8/2,3/1,8 µl | Plasmid DNA (P88/P89/P90) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl)/ PstI |
| required amount for total volume of 10 µl | ddH2O |
Ligation of F67 and F71, Transformation of E. coli XL1 blue afterwards
Investigator: Niklas
Aim of the experiment: Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 2,4 µl | Vektor |
| 7,6 µl | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
- next step: analytic digestion of transformation was successful
Digestion of PCR on genesynthesis 3 and 4, and pSB1C3
Investigator: Luisa
Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.
Procedure:
- Batches for analytical digestions:
| volume | reagent |
| 1 µl each | enzyme (SapI, HindIII, XbaI, AgeI) |
| 5 µl | CutSmart buffer (10x) |
| 41 µl | DNA (purified PCR-products of GSY3 and 4) |
- Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80.
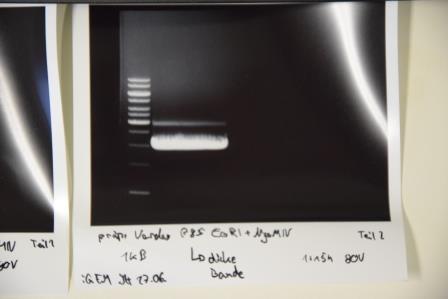
Analytical digestion and gelelectrophoresis of P80 , P78 and P85
Investigator: Julian
Aim of experiment: Analytical digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)
Procedure:
- Batches for analytical digestions:
- P80 and P78: EcoRI and AgeI
- P85: EcoRI and NgoMIV
| volume | reagent |
| 10 µl | Plasmid DNA |
| 32 µl | ddH2O |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme(10 U/µl)/ PstI |
| 50 µl | TOTAL |
Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into E. coli XL1 blue
Investigator: Luisa
Aim of the experiment: Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 4,3µl(for F75), 8,2µl (for F76) | Vector |
| 12,7µl (F75), 8,8 (F76) | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| =20 µl | TOTAL |
- Ligation was incubated at RT for 1,5h.
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator.
Chemical biotinylation of BSA
Investigator: Niklas
Procedure:
- BSA was chemically biotinylated with a 20x and 40x molar excess:
- 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85)
- dissolve BSA (10 mg/ml)
- Add biotin-NHS-ester: 20,5 mg for 40x molar excess
- reaction over night
Tuesday, June 28th
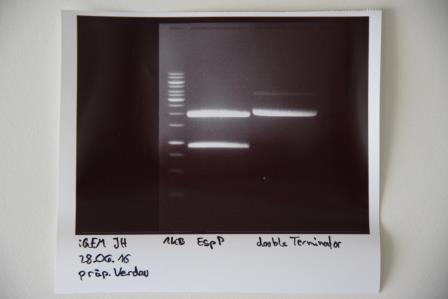
Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator)
Investigator: Julian
Aim of experiment: Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator)
Procedure:
- Batches for analytical digestions:
- P83: EcoRI and SpeI
- P29: EcoRI and XbaI
| volume | reagent |
| 6.5/13 µl | Plasmid DNA (P83/P29) |
| 35.5/29 µl | ddH2O (P83/P29) |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme |
| 50 µl | TOTAL |
- Preparative gelelectrophoresis was performed at 80 V for 80 min
Gelelectrophoresis of F73 (pSB1C3 digst. EcoRI, AgeI)
Investigator: Julian
Aim of experiment: gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)
Procedure: Preparative gelelectrophoresis was performed at 100 V for 45 min using a pocket spanning over the full width of the 1% agarose gel
Wednesday, June 29th
Transformation of F83 (EspP-Fragment in Double Terminator + pSB1C3), F84 (GSY3 + pSB1C3) F85 (GSY4 + pSB1C3)
Investigator: Niklas
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Cam-, Amp and Kan-plates
Result:
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Cam-, Amp and Kan-plates
Result:
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Cam-, Amp and Kan-plates
Result:
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Cam-, Amp and Kan-plates
Result:
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Cam-, Amp and Kan-plates
Result:
- After 12 hous of incubation there were some (~20) colonies on the 50µl-plate transformed with Cam-resistence-plasmids. The transformation seems to have been succesful, though cells grow slow.
- There were no colonies on the plates with the untransformed E.Coli. Hence we can assume that there are no other plasmids with resistences against kan, tet, amp or cam in our batch of competent cells.
Frisday, June 30th
Transformation of P9 (pASK with Streptavidin WT) and QC-PCR product into new E. coli XL1-blue
Aim of experiment: Tranforming pASK without NdeI-cutting point into E.coli (two batches from diffrent approches were tested). And Transforming pASK_SA1 into E.coli XL-1-blue.
Investigator: Luisa
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 30 sec. heat shock at 42 °C
- Adding of 950 µl LB-medium to each tube. 1h hour incubation at 37°C.
- The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C.