Parts
Our project required a system able to detect then chelate dietary copper. We decided that we should produce parts to test these two functions separately.
Having characterised each we could have a better understanding of a system whereby dietary copper was detected and a chelator produced until the free copper concentration is reduced to a stable level.
Our copper chelators belong in our list of basic parts whilst most of our promoters and promoter-chelators systems belong in our composite parts range. All can be found in our complete parts collection.
Copper chelators
In order to chelate copper we searched for copper binding and storage proteins.
We decided that the ideal copper chelator would have these properties:
- Should be able to bind multiple copper ions per peptide to increase the efficient use of cell resources.
- They should be from the prokaryotic domain because eukaryotic proteins can have expression issues in Escherichia coli.
- As E. coli naturally deals with copper toxicity by binding copper in the periplasm then exporting it, periplasmic proteins may reduce toxicity to the host.
Using these criteria we found two copper chelators that we thought would be useful. We designed gBlocks, codon optimised for E. coli, containing these parts both alone and linked to super-folder GFP. This form of the fluorescent protein was used because the standard GFP doesn’t fold particularly well in the periplasm where one of our chelators is intended to travel to.
The proteins, both alone and attached to sfGFP, were ordered with C terminal hexa-histidine tag so we could purify them. A concern was raised that the His tag would also weakly bind copper potentially affecting the results. However we decided that increasing the copper-binding would only improve the proteins' intended function.
Copper Storage Protein 1
Copper storage protein 1 is a protein discovered in a methane-oxidizing alphaproteobacterium called Methylosinus trichosporium OB3b. (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al.(1) discovered Csp1 in 2015, characterised the protein’s copper affinity and obtained crystal structures with and without copper.
TAT Csp1
Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins, which are unstructured until they fold around metal ion clusters. Vita et al.(1) found an average copper affinity of approximately 1x1017M-1.
Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely a periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity.
We codon optimised Csp1 to E. coli and replaced the original TAT sequence with a TAT sequence from the E. coli protein CueO, which is also involved in copper regulation. To get Csp1 from the shipping vector to the pBAD expression system for testing the TAT sequence had to be modified by the addition of a serine residue after the initiator methionine. Serine was chosen over other amino acid possibilities because other TAT sequences seemed to have serine in this location.
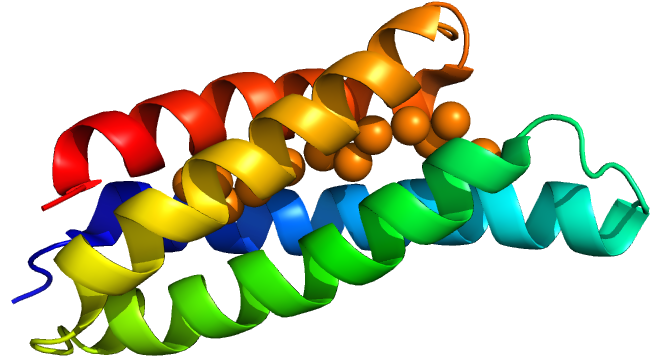
TAT Csp1 sfGFP
Csp1 sfGFP is a form of Csp1 with a C terminal sfGFP tag. The sfGFP has a C terminal hexahistidine tag for purifications. We used this version of Csp1 for purification and microscopy studies. The microscopy images unfortunately showed no evidence of the protein reaching the periplasm. Instead they likely formed inclusion bodies.
 Composite, Differential Interference Contrast and fluorescence channel images for Csp1sfGFP expressed form the pBad expression plasmid with 2mM arabinose
Composite, Differential Interference Contrast and fluorescence channel images for Csp1sfGFP expressed form the pBad expression plasmid with 2mM arabinose
Mycobacterial Metallothionien (MymT)
MymT is a small prokaryotic metallothein discovered in Mycobacterium tuberculosis by Gold et al(2). It is believed that the protein may help the bacterium survive copper toxicity. No crystal structure of MymT was obtained but there is an NMR structure of a eukaryotic homologue(3):

MymT
MymT is much smaller than Csp1 and, because we did not add in a secretion tag is intended to be cytoplasmic. It can bind up to 7 copper ions but has a preference for 4-6.(2)
MymT sfGFP
This was a form of MymT with a C terminal sfGFP. We used this version of MymT for purification and microscopy studies. Unlike Csp1sfGFP it had few problems with expression.
Promoters
For our copper chelating bacteria to successfully compete in the gut we investigated copper biosensors so that the chelator was only produced when copper was present, maintaining cell resources.
As copper is a redox metal it is both very important in cells enzymes but also very toxic because it leads to the formation of reactive oxygen species. Consequently Escherichia coli have two system of detecting excess copper: the CueR-linked system that detect cytoplasmic copper and the CusS/CusR two component system that detects periplasmic copper.
We investigated both systems and, by arranging the components to form a positive feedback loop hoped to improve their sensitivity over the range of copper concentrations we were interested in.
CueR linked systems
 The E. coli copper regulator CueR: monomer with copper (red) and without copper (cyan) attached to the pCopA promoter showing DNA bending
The E. coli copper regulator CueR: monomer with copper (red) and without copper (cyan) attached to the pCopA promoter showing DNA bending
E. coli cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase. More information and an animated version of the above image can be found here. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:
CCTTCCNNNNNNGGAAGG.
This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.(4)
We constructed different promoter systems based upon the promoter for copA, pCopA:
pCopA sfGFP

The simplest system we tested was simply the pCopA promoter in front sfGFP. We found that this system was only weakly copper responsive. This may be because there were 500+ copies of the pCopA promoter located on the high copy plasmid compared with only a single copy of CueR expressed from the bacterial genome. If CueR is only weakly expressed then there would be insufficient protein to bind at all of the promoter binding sites and the genes could not be activated.
A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location.

pCopA sfGFP with constitutively expressed CueR

The promoter system we ordered was based upon this part in the registry from Team Bielefeld-CeBiTec in 2015.
The team assembled a copper biosensor from two subparts which they then joined together:

The first part is a pCopA-RBS-sfGFP and the second part is the regulator CueR expressed from a constitutive promoter.
However the part sequence they deposited differs from the diagram as shown above as the constitutive promoter-CueR part has been joined to the other part the wrong way round and labelled as if it were correct.
The sequence as it has been deposited has the constitutive promoter controlling CueR on the same strand as pCopA and sfGFP and facing in the same direction.
As they are not separated by a transcriptional terminator, transcription beginning from the constitutive promoter could produce mRNA for both CueR and sfGFP.
When we designed this part we flipped the CueR and constitutive promoter to face the opposite direction on the opposite strand. We also had to remove the 5'UTR because it was too AT rich to be synthesised.
Unfortunately every attempt to amplify this part from the synthesised sequence we received from IDT resulted in the same two point mutations in the sfGFP region of this part making it non-functional. To compare this promoter system to the other we designed we used our parts with chelator-sfGFP fusions instead of the sfGFP which we expected to have similar behaviour.
pCopA with constitutively expressed CueR
To facilitate making any protein under control of a copper-responsive promoter we designed a part with similar to the above pCopA sfGFP with constitutively expressed CueR but without the sfGFP. The RBS however is still present meaning that any protein with the ATG biobrick prefix and universal suffix can be inserted with biobrick standard assembly with the correct RBS-ATG distance.
pCopA CueR sfGFP/ Feedback pCopA sfGFP

We designed a part similar to the above but with CueR expressed from the same pCopA promoter that it controls in order to study CueR operating under a feedback loop. If CueR is acting as a net activator, this should in theory act as a positive feedback system whereby a small amount of copper stimulation produces more CueR causing greater activation. Should CueR be acting as a net repressor this system would be less sensitive than our previous system and this appears to be what we discovered.
A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location.

CusS/CusR linked systems
The second system E. coli uses to respond to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator CusR.
When CusS binds periplasmic copper it transfer a phosphate group from ATP to CusR aspartate residue 51 via CusS histidine residue 271. Phosphorylated CusR can bind to DNA inverted repeat CusR boxes (AAAATGACAANNTTGTCATTTT) and activate gene expression.
In E. coli this box is present between the promoters for CusCFBA operon which encodes a multi-protein pump that exports cytoplasmic and periplasmic copper from the cell and the CusRS operon which encodes the two component system (therefore acting as a positive feedback loop in vivo). (4)
pCusC RFP

We received a copy of pCusC mKate (a form of RFP) from Tom Folliard, one of the PhD students in our lab. This sequence had a biobrick-illegal Spe1 site in the RBS region meaning we could not deposit in the registry. We performed site directed mutagenesis to swap a C in the Spe1 site for G, and moved it into the shipping vector. This part was then deposited. We also amplified the promoter region only and deposited this separately.

pCusC CusR RFP

We also designed a version of the part with CusR expressed before mKate forming a positive feedback loop. This sort of system was shown to be more responsive in a paper by Ravikumar S et al.(5). However we were unable to clone a form of the part with a point mutation which appears to have broken the feedback loop:
A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location.

Composite Parts
These are parts we produced with a copper chelator under control of a copper-sensitive promoter. If these behave as expected then, if a small concentration of copper is added to cells containing this part, copper will induce expression of the chelator, which would then chelate free copper. If free copper is reduced then chelator expression should be reduced until a stable equilibrium is reached. This copper buffering is dependent upon both the copper promoters behaviour and the copper chelator expression and affinity (see our modelling page). In practice the level of copper concentrations upon which these systems would operate was below our copper absorbance assay detection limits.
pCopA MymT with constitutively expressed CueR

This part contains the chelator MymT behind a copper sensitive promoter.
pCopA MymT sfGFP constitutively expressed CueR

This part contains our chelator MymT with a C terminal sfGFP tag behind the pCopA promoter with constitutively expressed CueR.

pCopA TAT Csp1 sfGFP constitutively expressed CueR

This part contains our chelator Csp1 with a C terminal sfGFP tag behind the pCopA promoter with constitutively expressed CueR. Likely due to the expression problems of Csp1 it responded worse than the similar part above with our MymTsfGFP fusion protein.

References:
-
(1) Nicolas Vita, Semeli Platsaki, Arnaud Basle, Stephen J. Allen, Neil G. Paterson, Andrew T. Crombie, J. Colin Murrell, Kevin J.Waldron & Christopher Dennison (2015)
“A four-helix bundle stores copper for methane
oxidation”, Nature 525 issue 7567 pg. 140-143 doi:10.1038/nature14854
-
(2) Ben Gold, Haiteng Deng, Ruslana Bryk, Diana Vargas, David Eliezer, Julia Roberts,
Xiuju Jiang, & Carl Nathan (2009) “Identification of a Copper-Binding Metallothionein in Pathogenic Mycobacteria” Nat Chem Biol.
2008 October ; 4(10): 609–616. doi:10.1038/nchembio.109.
-
(3)
1AQS from Peterson, C.W., Narula, S.S., Armitage, I.M. “3D solution structure of copper and silver-substituted yeast metallothioneins” (1996) FEBS Lett. 379: 85-93
-
(4) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27.
-
(5) Sambandam Ravikumar, Van Dung Pham, Seung Hwan Lee, Ik-keun Yoo, Soon Ho Hong (2012) “Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop” Journal of Industrial Microbiology & Biotechnology June 2012, Volume 39, Issue 6, pp 861–868




















