| (9 intermediate revisions by the same user not shown) | |||
| Line 93: | Line 93: | ||
</p> | </p> | ||
<p> | <p> | ||
| − | Having characterised each we could have a better understanding of a system whereby dietary copper was detected and a <a data-toggle="popover1" data-trigger="hover" placement:"top" title="Chelator" data-content="A molecule, here a protein, able to | + | Having characterised each we could have a better understanding of a system whereby dietary copper was detected and a <a data-toggle="popover1" data-trigger="hover" placement:"top" title="Chelator" data-content="A molecule, here a protein, able to form multiple bonds to a single metal ion.">chelator</a> produced until the free copper concentration is reduced to a stable level. |
</p> | </p> | ||
<p> | <p> | ||
| Line 138: | Line 138: | ||
<h2 id="MymTH">Mycobacterial Metallothionien (MymT)</h2> | <h2 id="MymTH">Mycobacterial Metallothionien (MymT)</h2> | ||
<p> | <p> | ||
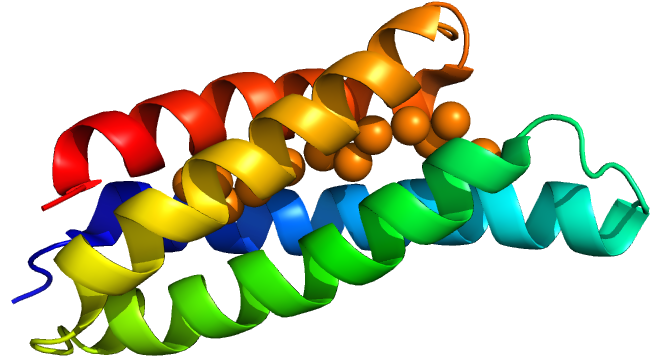
| − | MymT is a small prokaryotic <a data-toggle="popover1" data-trigger="hover" title="Metallothein" data-content="Very small Cysteine-rich proteins involved in transport and storage">metallothein</a> discovered in <i>Mycobacterium tuberculosis</i> by Gold <i>et al.</i><sup>(2)</sup> | + | MymT is a small prokaryotic <a data-toggle="popover1" data-trigger="hover" title="Metallothein" data-content="Very small Cysteine-rich proteins involved in transport and storage">metallothein</a> discovered in <i>Mycobacterium tuberculosis</i> by Gold <i>et al.</i><sup>(2)</sup> It is believed that the protein may help the bacterium survive copper toxicity. No crystal structure of MymT was obtained but there is an NMR structure of a eukaryotic homologue<sup>(3)</sup>: |
</p> | </p> | ||
<img src="https://static.igem.org/mediawiki/2016/c/ca/MymT_image_for_parts_page_Sam_Oxford_2016.png" width="25%" /><figcaption>NMR average of yeast Cu Metallothein (PDB: 1AQR)</figcaption> | <img src="https://static.igem.org/mediawiki/2016/c/ca/MymT_image_for_parts_page_Sam_Oxford_2016.png" width="25%" /><figcaption>NMR average of yeast Cu Metallothein (PDB: 1AQR)</figcaption> | ||
| Line 144: | Line 144: | ||
<h3 id="MymT">MymT</h3> | <h3 id="MymT">MymT</h3> | ||
<p> | <p> | ||
| − | MymT is much smaller than Csp1 and, because we did not add in a secretion tag is intended to be cytoplasmic. It can bind up to 7 copper ions but has a preference for 4-6.<sup>(2)</sup> | + | MymT is much smaller than Csp1 and, because we did not add in a secretion tag, is intended to be cytoplasmic. It can bind up to 7 copper ions but has a preference for 4-6.<sup>(2)</sup> |
</p> | </p> | ||
| Line 168: | Line 168: | ||
<img src="https://static.igem.org/mediawiki/2016/4/47/T--Oxford--cueRtoDNA.png" width="25%" /><figcaption>The <i>E. coli</i> copper regulator CueR: monomer with copper (red) and without copper (cyan) attached to the pCopA promoter showing DNA bending. (cealigned in pymol from PDB 4wls and 4wlw)<sup>(6)</sup></figcaption> | <img src="https://static.igem.org/mediawiki/2016/4/47/T--Oxford--cueRtoDNA.png" width="25%" /><figcaption>The <i>E. coli</i> copper regulator CueR: monomer with copper (red) and without copper (cyan) attached to the pCopA promoter showing DNA bending. (cealigned in pymol from PDB 4wls and 4wlw)<sup>(6)</sup></figcaption> | ||
<p> | <p> | ||
| − | <i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a <a data-toggle="popover1" data-trigger="hover" title="MerR" data-content="An E. coli transcription factor with a helix-turn-helix motif that regulates the bacterial cells’ response to mercury ">MerR</a>-type regulator with an interesting <a href="https://2016.igem.org/Team:Oxford/CueR_MOI">mechanism of action</a> whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with <a data-toggle="popover1" data-trigger="hover" title="RNA polymerase" data-content="An enzyme that transcribes the DNA sequence of a gene into an mRNA sequence that is then translated into a protein sequence.">RNA polymerase</a><sup>(7)</sup> | + | <i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a <a data-toggle="popover1" data-trigger="hover" title="MerR" data-content="An E. coli transcription factor with a helix-turn-helix motif that regulates the bacterial cells’ response to mercury ">MerR</a>-type regulator with an interesting <a href="https://2016.igem.org/Team:Oxford/CueR_MOI">mechanism of action</a> whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with <a data-toggle="popover1" data-trigger="hover" title="RNA polymerase" data-content="An enzyme that transcribes the DNA sequence of a gene into an mRNA sequence that is then translated into a protein sequence.">RNA polymerase</a>.<sup>(7)</sup> More information and an animated version of the above image can be found <a href="https://2016.igem.org/Team:Oxford/CueR_MOI">here</a>. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA <a data-toggle="popover1" data-trigger="hover" title="Inverted Repeat" data-content="A DNA sequence followed downstream by its reverse complement">inverted repeats</a> called CueR boxes with the sequence: |
</p> | </p> | ||
<p> | <p> | ||
| Line 190: | Line 190: | ||
<p>A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | <p>A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/f/fd/Pcg_graph_4h_sam_oxford_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/f/fd/Pcg_graph_4h_sam_oxford_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980005 after 4 hours </figcaption> |
| − | <h3 id="pg">pCopA sfGFP with divergent | + | <h3 id="pg">pCopA sfGFP with divergent CueR</h3> |
<img src="https://static.igem.org/mediawiki/2016/6/6d/Pg_diagram_v1_Sam_Oxford_2016.jpeg" width="50%" /><figcaption>Our biosensor design. We unfortunately never managed to submit this part</a></figcaption> | <img src="https://static.igem.org/mediawiki/2016/6/6d/Pg_diagram_v1_Sam_Oxford_2016.jpeg" width="50%" /><figcaption>Our biosensor design. We unfortunately never managed to submit this part</a></figcaption> | ||
<p> | <p> | ||
| Line 216: | Line 216: | ||
<p> | <p> | ||
When we designed this part we flipped the CueR and the constitutive promoter to face the opposite direction on the opposite strand i.e. so they were divergent. We also had to remove the 5'UTR, which Bielefeld found to increase expression, because it was too AT rich to be synthesised.</p> | When we designed this part we flipped the CueR and the constitutive promoter to face the opposite direction on the opposite strand i.e. so they were divergent. We also had to remove the 5'UTR, which Bielefeld found to increase expression, because it was too AT rich to be synthesised.</p> | ||
| − | <p> Unfortunately, every attempt to amplify this part from the synthesised sequence we received from IDT resulted in the same two point mutations in the sfGFP region of this part making it non-functional. To compare this promoter system to the others we designed | + | <p> Unfortunately, every attempt to amplify this part from the synthesised sequence we received from IDT resulted in the same two point mutations in the sfGFP region of this part making it non-functional. To compare this promoter system to the others we designed, we used our parts with chelator-sfGFP fusions instead of the sfGFP which we expected to have similar behaviour. |
</p> | </p> | ||
<h3 id="p">pCopA with divergent expressed CueR</h3> | <h3 id="p">pCopA with divergent expressed CueR</h3> | ||
<p> | <p> | ||
| − | To facilitate making any protein under control of a copper-responsive promoter we designed a part (<a href="http://parts.igem.org/Part:BBa_K1980006">BBa_K1980006</a>) | + | To facilitate making any protein under control of a copper-responsive promoter we designed a part (<a href="http://parts.igem.org/Part:BBa_K1980006">BBa_K1980006</a>) similar to the above pCopA sfGFP with divergent expressed CueR but without the sfGFP. The RBS however is still present meaning that any protein with the ATG biobrick prefix and universal suffix can be inserted with biobrick standard assembly with the correct RBS-ATG distance. |
</p> | </p> | ||
| Line 231: | Line 231: | ||
A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/9/99/Fcg_4h_graph_sam_oxford_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/9/99/Fcg_4h_graph_sam_oxford_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980008 after 4 hours </figcaption> |
<h2 id="CusS-CusR">CusS/CusR linked systems</h2> | <h2 id="CusS-CusR">CusS/CusR linked systems</h2> | ||
<p> | <p> | ||
| − | The second system <i>E. coli</i> uses to respond to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator CusR. | + | The second system <i>E. coli</i> uses to respond to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme: CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator: CusR. |
</p> | </p> | ||
<p> | <p> | ||
| Line 244: | Line 244: | ||
<h3 id="pCusC">pCusC RFP</h3> | <h3 id="pCusC">pCusC RFP</h3> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/1/1a/PCusC_mKate_diagram_v1_Sam_Oxford_2016.jpeg" width="50%" /><figcaption>The design of part <a href="http://parts.igem.org/Part: | + | <img src="https://static.igem.org/mediawiki/2016/1/1a/PCusC_mKate_diagram_v1_Sam_Oxford_2016.jpeg" width="50%" /><figcaption>The design of part <a href="http://parts.igem.org/Part:BBa_K1980007">BBa_K1980007</a></figcaption> |
<p> | <p> | ||
We received a copy of pCusC <a data-toggle="popover1" data-trigger="hover" title="monomeric Kate2" data-content="A very bright far-red fluorescent protein with excitation/emission maxima at 588 and 633 nm">mKate</a> (a form of RFP) from Tom Folliard, one of the PhD students in our lab. This sequence had a biobrick-illegal Spe1 site in the RBS region meaning we could not deposit in the registry. We performed site directed mutagenesis to swap a C in the Spe1 site for G, and moved it into the shipping vector. This part was then deposited. We also amplified the promoter region only and deposited this separately.(<a href="http://parts.igem.org/Part:BBa_K1980004">BBa_K1980004</a>) | We received a copy of pCusC <a data-toggle="popover1" data-trigger="hover" title="monomeric Kate2" data-content="A very bright far-red fluorescent protein with excitation/emission maxima at 588 and 633 nm">mKate</a> (a form of RFP) from Tom Folliard, one of the PhD students in our lab. This sequence had a biobrick-illegal Spe1 site in the RBS region meaning we could not deposit in the registry. We performed site directed mutagenesis to swap a C in the Spe1 site for G, and moved it into the shipping vector. This part was then deposited. We also amplified the promoter region only and deposited this separately.(<a href="http://parts.igem.org/Part:BBa_K1980004">BBa_K1980004</a>) | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/9/92/PCusC_RFP_4h_graph_Oxford_Sam_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/9/92/PCusC_RFP_4h_graph_Oxford_Sam_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980007 after 4 hours </figcaption> |
<h3 id="fck">pCusC CusR RFP</h3> | <h3 id="fck">pCusC CusR RFP</h3> | ||
| Line 255: | Line 255: | ||
We also designed a version of the part with CusR expressed before mKate forming a positive feedback loop. This sort of system was shown to be more responsive in a paper by Ravikumar S <i>et al.</i><sup>(5)</sup>. However after many attempts at cloning our only full length construct had a point mutation (Val to Ala) in the <i>CusR</i> gene. We tested this part to see if this mutation was tolerable, but found no evidence for a more sensitive system: | We also designed a version of the part with CusR expressed before mKate forming a positive feedback loop. This sort of system was shown to be more responsive in a paper by Ravikumar S <i>et al.</i><sup>(5)</sup>. However after many attempts at cloning our only full length construct had a point mutation (Val to Ala) in the <i>CusR</i> gene. We tested this part to see if this mutation was tolerable, but found no evidence for a more sensitive system: | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/b/b7/Fck_4h_graph_sam_oxford_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/b/b7/Fck_4h_graph_sam_oxford_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980009 after 4 hours </figcaption> |
<p>A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | <p>A BspH1 site was included at the start codon of the sfGFP so that other proteins with a compatible start (such as the copper chelators we ligated into our pBAD vector) could be ligated into this location. | ||
</p> | </p> | ||
| Line 272: | Line 272: | ||
</p> | </p> | ||
| − | <h3 id="pmg">pCopA MymT sfGFP divergent expressed CueR</h3 | + | <h3 id="pmg">pCopA MymT sfGFP divergent expressed CueR</h3> |
| − | <img src="https://static.igem.org/mediawiki/2016/9/90/Pmg_diagram_v1_Oxford_sam_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/9/90/Pmg_diagram_v1_Oxford_sam_2016.jpeg" width="50%" /><figcaption>The design of part <a href="http://parts.igem.org/Part:BBa_K1980012">BBa_K1980012</a></figcaption> |
<p> | <p> | ||
This part contains our chelator MymT with a C-terminal sfGFP tag behind the pCopA promoter with divergent expressed CueR in the biobrick. | This part contains our chelator MymT with a C-terminal sfGFP tag behind the pCopA promoter with divergent expressed CueR in the biobrick. | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/4/42/Pmg_4h_graph_sam_oxofrd_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/4/42/Pmg_4h_graph_sam_oxofrd_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980012 after 4 hours </figcaption> |
<h3 id="ptcg">pCopA TAT Csp1 sfGFP constitutively expressed CueR</h3> | <h3 id="ptcg">pCopA TAT Csp1 sfGFP constitutively expressed CueR</h3> | ||
| Line 284: | Line 284: | ||
This part contains our chelator Csp1 with a C-terminal sfGFP tag behind the pCopA promoter with constitutively expressed CueR in the biobrick. Likely due to the expression problems of Csp1 it responded worse than the similar part above with our MymTsfGFP fusion protein. | This part contains our chelator Csp1 with a C-terminal sfGFP tag behind the pCopA promoter with constitutively expressed CueR in the biobrick. Likely due to the expression problems of Csp1 it responded worse than the similar part above with our MymTsfGFP fusion protein. | ||
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/9/97/Ptcg_graph_4h_sam_oxford_2016.jpeg" width="50%" /> | + | <img src="https://static.igem.org/mediawiki/2016/9/97/Ptcg_graph_4h_sam_oxford_2016.jpeg" width="50%" /><figcaption>Plate reader data for BBa_K1980010 after 4 hours </figcaption> |
</section> | </section> | ||
Latest revision as of 16:53, 3 November 2016