| (27 intermediate revisions by 5 users not shown) | |||
| Line 50: | Line 50: | ||
<div class="col-md-9 content-right" style="background-color: #fff;"> | <div class="col-md-9 content-right" style="background-color: #fff;"> | ||
| − | <div class="pageTitle pageTitleOrange"> | + | <div class="pageTitle pageTitleOrange">Cure</div> |
| + | <p><center><font size="5"> Our project was awarded a <font color="#FFD700"><b>GOLD</b></font> medal at the 2016 Jamboree. <br> We were nominated for: <b>Best Therapeutic Project</b>, | ||
| + | <br> | ||
| + | <b>Best Wiki</b>, | ||
| + | <b>Best Presentation</b> and <br> | ||
| + | <b>Best Education & Public Engagement!</b></center></font></p> | ||
<section id="Overview"> | <section id="Overview"> | ||
<h2>Overview</h2> | <h2>Overview</h2> | ||
<p> | <p> | ||
| − | Rare, or “orphan”, diseases are frequently ignored by the pharmaceutical industry. They encompass a huge range of disorders | + | Rare, or “orphan”, diseases are frequently ignored by the pharmaceutical industry. They encompass a huge range of disorders from ALS to Tourette’s Syndrome, but individually have a relatively low number of patients. The low patient numbers mean that there is very little impetus for the pharmaceutical industry to research and produce novel, innovative therapeutics. This means that patients are often left with unsatisfactory treatments. Our goal is to produce a probiotic therapeutic to treat one such disorder: Wilson’s Disease. |
</p> | </p> | ||
</section> | </section> | ||
| Line 60: | Line 65: | ||
<h2>Wilson's Disease</h2> | <h2>Wilson's Disease</h2> | ||
<p> | <p> | ||
| − | Wilson’s Disease is a genetic disorder characterised by an inability of the body to | + | Wilson’s Disease is a genetic disorder characterised by an inability of the body to metabolise copper. Normally, when copper is ingested, it is taken up by the small intestine, taken to the liver and subsequently transported into the blood or excreted into the bile. In Wilson’s Disease, there is a mutation in the gene ATP7B. ATP7B encodes a copper-transporting protein that is responsible for loading copper onto caeruloplasmin for transport in the blood, and into the bile for removal from the body. In the absence of a functional form of this protein, copper is unable to be removed from the liver after absorption. This results in toxic accumulation, as cuprous ions react with hydrogen peroxide to produce dangerous free radicals that damage tissue. This allows copper ions to leak into the blood and eventually accumulate in, and damage, other tissues, such as the kidneys and brain. You can read more about Wilson's disease <a href="https://2016.igem.org/Team:Oxford/Wilson_Disease">here</a>. |
</p><p> | </p><p> | ||
| − | Current treatments are regarded by patients as unsatisfactory. From discussions with | + | Current treatments are regarded by patients as unsatisfactory. From discussions with patients, we have identified three main problems with current treatments: |
</p> | </p> | ||
| − | <li class="textLiFont">Side effects: sometimes | + | <li class="textLiFont">Side effects: sometimes so severe that treatment must be stopped</li> |
| − | <li class="textLiFont">Price: these drugs are very expensive for the | + | <li class="textLiFont">Price: these drugs are very expensive for the UK National Health Service</li> |
| − | <li class="textLiFont">High | + | <li class="textLiFont">High dosage frequency: drugs must be taken daily and refrigerated</li> |
<p> | <p> | ||
| − | Our | + | Our Cure aims to address these limitations.</a> |
</p> | </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2016/d/d5/Kayser-fleischer_ring_sam_oxford_2016.jpeg" width="50%" style="border-radius: 20px"/><figcaption>Wilson's disease causes distinctive copper | + | <img src="https://static.igem.org/mediawiki/2016/d/d5/Kayser-fleischer_ring_sam_oxford_2016.jpeg" width="50%" style="border-radius: 20px"/><figcaption>Wilson's disease causes distinctive copper deposits in the cornea of some patients, causing brown rings around the iris called Kayser-Fleischer rings.</figcaption> |
</section> | </section> | ||
<section id="Probiotics"><br><br> | <section id="Probiotics"><br><br> | ||
<h2>Probiotics</h2> | <h2>Probiotics</h2> | ||
<p> | <p> | ||
| − | A probiotic | + | A probiotic is a microorganism that is introduced into the body for its beneficial properties. The concept of a probiotic, meaning “for life”, was introduced by Elie Metchnikoff in 1907, when he hypothesised that replacing or diminishing the populations of ‘putrefactive’ bacteria in the gut with lactic acid bacteria could positively affect bowel health. |
</p> | </p> | ||
<p> | <p> | ||
| − | Products that are commonly sold as probiotics include | + | Products that are commonly sold as probiotics include foodstuffs, such as yoghurts and cheeses. However, recently there has been an increase in the amount of research going into the use of probiotics as therapeutics, with the genetic engineering of organisms to produce useful substances. Currently there is limited legislation regarding probiotics, as probiotics sold as dietary supplements do not require FDA approval. A genetically-engineered probiotic therapeutic would require more stringent legislation and FDA approval to ensure, through clinical trials, that it works as expected. |
</p> | </p> | ||
<p> | <p> | ||
| − | Although advancing rapidly, the field of probiotics still requires significant research particularly in areas such as safety. Although regarded as safe for relatively healthy humans to consume, there have been some reports of probiotic-related side effects in people with serious underlying | + | Although advancing rapidly, the field of probiotics still requires significant research particularly in areas such as safety. Although regarded as safe for relatively healthy humans to consume, there have been some reports of probiotic-related side effects in people with serious underlying medical conditions. We carried out a comprehensive <a href="https://2016.igem.org/Team:Oxford/Safety">safety</a> review when completing our project. |
</p> | </p> | ||
</section> | </section> | ||
| Line 88: | Line 93: | ||
<h2>Video</h2> | <h2>Video</h2> | ||
<video controls style="width:100%; border:1px solid #3F3F3F;"> | <video controls style="width:100%; border:1px solid #3F3F3F;"> | ||
| − | <source src="https://static.igem.org/mediawiki/2016/ | + | <source src="https://static.igem.org/mediawiki/2016/e/e0/T--Oxford--finalvideofinal.mp4" type="video/mp4"/> |
</video> | </video> | ||
</section> | </section> | ||
| Line 95: | Line 100: | ||
<h2>Parts</h2> | <h2>Parts</h2> | ||
<p> | <p> | ||
| − | In order to detect and | + | In order to provide a long-term treatment for Wilson's disease we realised that we needed to design components of a bacterial system that could detect copper and produce a copper chelator to prevent its absorption by the body. |
</p> | </p> | ||
| + | <p> | ||
| + | We investigated copper detection using promoter systems based upon the native <i>E. coli</i> copper regulator <a href="https://2016.igem.org/Team:Oxford/Parts#CueR">CueR</a> and the <a href="https://2016.igem.org/Team:Oxford/Parts#CusS-CusR">CusS/CusR</a> two component system. The two copper chelators we tested were <a href="https://2016.igem.org/Team:Oxford/Parts#Csp1H">Copper storage protein 1</a> (Csp1) from <i>Methylosinus trichosporium</i> OB3b and <a href="https://2016.igem.org/Team:Oxford/Parts#MymTH">mycobacterial metallothionein</a> (MymT) from <i>Mycobacterium tuberculosis.</i></p> | ||
| + | <p>You can read about how we chose our parts <a href="https://2016.igem.org/Team:Oxford/Parts">here</a>.</p> | ||
<img src="https://static.igem.org/mediawiki/2016/0/0e/Csp1_single_for_Chelators_page_Sam_Oxford_2016.png" width="50%"/><figcaption>Our chelator Copper Storage Protein 1 (monomer) with 13 bound copper ions.</figcaption> | <img src="https://static.igem.org/mediawiki/2016/0/0e/Csp1_single_for_Chelators_page_Sam_Oxford_2016.png" width="50%"/><figcaption>Our chelator Copper Storage Protein 1 (monomer) with 13 bound copper ions.</figcaption> | ||
</section> | </section> | ||
| Line 110: | Line 118: | ||
<h2>Results</h2> | <h2>Results</h2> | ||
<p> | <p> | ||
| − | Through our experimental work we have been able to obtain data suggesting the | + | Through our experimental work we have been able to obtain data suggesting the following:</p> |
<ul> | <ul> | ||
<li>Different arrangements of both our CueR-linked and CusS/CusR-linked promoter systems are sensitive over a range of copper concentrations, including at the lower concentrations mimicking the gut.</li> | <li>Different arrangements of both our CueR-linked and CusS/CusR-linked promoter systems are sensitive over a range of copper concentrations, including at the lower concentrations mimicking the gut.</li> | ||
<li>MymT is able to chelate a measurable amount of copper <i>in vivo.</i></li> | <li>MymT is able to chelate a measurable amount of copper <i>in vivo.</i></li> | ||
| − | <li>Alternately layered chitosan-alginate beads release material in the small intestine, following passage through the stomach. </li> | + | <li>Alternately layered chitosan-alginate beads release material in the small intestine, following passage through the stomach, in a simulation accounting for the pH change. </li> |
</ul> | </ul> | ||
<p>Please see our <a href="https://2016.igem.org/Team:Oxford/Experiments">experiments</a> and <a href="https://2016.igem.org/Team:Oxford/Results">results</a> pages for more discussion of our methods and the outcomes.</p> | <p>Please see our <a href="https://2016.igem.org/Team:Oxford/Experiments">experiments</a> and <a href="https://2016.igem.org/Team:Oxford/Results">results</a> pages for more discussion of our methods and the outcomes.</p> | ||
| Line 123: | Line 131: | ||
<h2>Improving Registry Parts</h2> | <h2>Improving Registry Parts</h2> | ||
<p> | <p> | ||
| − | As part of our project, we have improved the function/characterisation of three poorly | + | As part of our project, we have improved the function/characterisation of three poorly documented parts.</p> |
<p> | <p> | ||
Please visit our <a href="https://2016.igem.org/Team:Oxford/Parts">parts</a> and <a href="https://2016.igem.org/Team:Oxford/Results">results</a> pages for further information.</p> | Please visit our <a href="https://2016.igem.org/Team:Oxford/Parts">parts</a> and <a href="https://2016.igem.org/Team:Oxford/Results">results</a> pages for further information.</p> | ||
<h3>BBa_I760005:</h3> | <h3>BBa_I760005:</h3> | ||
| − | <p> | + | <p>This <a href="http://parts.igem.org/Part:BBa_I760005">copy of pCusC</a> located in the registry had been previously used 11 times, but had no associated useful characterisation data. From discussions with a mentor, Tom Folliard, we came to the conclusion that this may have been because the promoter region was too short, and thus missing key binding sites. We decided to deposit an elongated form of this promoter, which we have achieved thorough characterisation. </p> |
<img src="https://static.igem.org/mediawiki/2016/9/92/PCusC_RFP_4h_graph_Oxford_Sam_2016.jpeg" width="50%" /><figcaption>Plate reader data for our improved part (BBa_K1980004) with RFP: BBa_K1980007</figcaption> | <img src="https://static.igem.org/mediawiki/2016/9/92/PCusC_RFP_4h_graph_Oxford_Sam_2016.jpeg" width="50%" /><figcaption>Plate reader data for our improved part (BBa_K1980004) with RFP: BBa_K1980007</figcaption> | ||
<h3>BBa_K190020:</h3> | <h3>BBa_K190020:</h3> | ||
| − | <p>MymT can be found in the registry with no associated characterisation data. We codon-optimised the sequence for expression in <i>E. coli</i> and added a hexahistidine tag for purification. We also produced a form with a C-terminal sfGFP tag | + | <p><a href="http://parts.igem.org/Part:BBa_K190020">MymT</a> can be found in the registry with no associated characterisation data. We codon-optimised the sequence for expression in <i>E. coli</i> and added a hexahistidine tag for purification. We also produced a form with a C-terminal sfGFP tag (also his-tagged) to aid purification, allowing us to view the cellular distribution of MymT and allow analysis by <u>F</u>luorescence <u>L</u>ifetime <u>IM</u>aging (FLIM). The fluorescence lifetime of GFP with a C-terminal His tag was shown to be reduced by copper by Hötzer et al., allowing FLIM to be used in an <i>in vivo</i> assay of copper binding. This has provided us with preliminary characterisation data for the protein, suggesting copper-binding activity <i>in vivo</i>. More details can be found on our <a href="https://2016.igem.org/Team:Oxford/Results#FLIM">results page</a>. </p> |
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/7/7e/New_flim_plot_image_sam_oxford_2016.png" width="50%" /><figcaption>Fluorescence lifetime for sfGFP, Csp1-sfGFP and MymTsfGFP from iGEM Cardiff. (No error bars shown when standard deviation=0.) </figcaption> |
<h3>BBa_K1758324:</h3> | <h3>BBa_K1758324:</h3> | ||
| − | <p> | + | <p>We were interested in using <a href="http://parts.igem.org/Part:BBa_K1758324">this part</a> from Team Bielefeld-CeBiTec in 2015 as a potential copper biosensor. |
| + | <p> | ||
| + | The team assembled a copper biosensor from two subparts which they then joined together: | ||
| + | </p> | ||
| + | <p> | ||
| + | The first part is a pCopA-RBS-sfGFP and the second part is the regulator CueR expressed from a constitutive promoter. The part is deposited in the registry and labelled to suggest the CueR is expressed divergent from the sfGFP (on the opposite strand and transcribed in the opposite direction): | ||
| + | </p> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/2/20/PG_incorrect_Oxford_Sam_2016.png" width="75%" /><figcaption>How BBa_K1758324 is labelled in the registry</figcaption> | ||
| + | <p> | ||
| + | However if you look at the sequence level this is clearly not the case. The constitutive promoter and the CueR start codon are at the 5’ end of the sfGFP coding strand and the CueR stop codon just upstream of pCopA. The part in fact has the constitutive promoter on the same strand as pCopA and sfGFP facing in the same direction and would be better represented like this: | ||
| + | </p> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/0/09/PCopA_CueR_incorrect_LOL_sam_oxford_2016.png" width="70%" /><figcaption>How BBa_K1758324 should be labelled in the registry based on its underlying sequence</figcaption> | ||
| + | <p> | ||
| + | As the two coding regions are not separated by a transcription terminator, there would be read through from the constitutive promoter to the sfGFP and sfGFP would be expressed even in the absence of copper. As no negative control is included in the plate reader graph they provide and no settings provided for their BioLector experiments in their <a href="https://2015.igem.org/Team:Bielefeld-CeBiTec/Protocols">protocols</a> it is unclear just how high the expression level at 0mM copper was for this part compared to a negative control strain. | ||
| + | </p> | ||
| + | <p>The CueR subpart (<a href="http://parts.igem.org/Part:BBa_K17583204">BBa_K1758320</a>) making up BBa_K1758324 is also incorrectly labelled.</p> | ||
| + | <p> | ||
| + | When we designed this part we flipped the CueR and the constitutive promoter to face the opposite direction on the opposite strand i.e. so they were divergent. We also had to remove the 5'UTR, which Bielefeld found to increase expression, because it was too AT rich to be synthesised.</p> | ||
| + | <p> Unfortunately, every attempt to amplify this part from the synthesised sequence we received from IDT resulted in the same two point mutations in the sfGFP region of this part making it non-functional. To compare this promoter system to the others we designed, we used our parts with chelator-sfGFP fusions instead of the sfGFP which we expected to have similar behaviour. | ||
| + | </p></p> | ||
</section> | </section> | ||
<section id="Conc"> | <section id="Conc"> | ||
<h2>Conclusion</h2> | <h2>Conclusion</h2> | ||
| − | <p>Over the course of the summer we have successfully created and submitted 13 sequence-confirmed BioBrick parts, 8 of which have been characterised. Through our | + | <p>Over the course of the summer we have successfully created and submitted 13 sequence-confirmed BioBrick parts, 8 of which have been characterised. Through our experiments, we have been able to examine the copper-sensitivity of a variety of promoters integral to our Cure. In addition, we have tested the chelating-ability of our copper chelators with a variety of different assays. Although these have not all been successful, through a collaboration with Cardiff we have been able to obtain preliminary data suggesting that MymT may be able to successfully lower the intracellular copper concentration. We have also obtained promising preliminary data relating to the delivery of substances to the small intestine via alginate-chitosan beads.</p> |
| − | <p>In conclusion, | + | <p>In conclusion, over the course of the summer we have successfully managed to produce an initial proof of concept for our idea and have contributed to our understanding of these systems, in addition to investigating a treatment that addresses the needs and concerns of the patients themselves.</p> |
</section> | </section> | ||
| Line 147: | Line 174: | ||
<p>To further develop our project, we would hope to carry out a number of further experiments to allow us to generate something more than a proof-of-concept model. Primarily, we would hope to properly characterise the copper-chelating ability of Csp1 and confirm its localisation in the periplasm by redesigning the TAT sequence. Additionally, we would carry out further experiments to refine our delivery system. </p> | <p>To further develop our project, we would hope to carry out a number of further experiments to allow us to generate something more than a proof-of-concept model. Primarily, we would hope to properly characterise the copper-chelating ability of Csp1 and confirm its localisation in the periplasm by redesigning the TAT sequence. Additionally, we would carry out further experiments to refine our delivery system. </p> | ||
<p>Once the system was able to successfully operate in <i>E. coli</i> DH5-α, we would hope to express it in a strain that more closely mimics the bacteria of the gut. More information on a potential future chassis choice can be found on our <a href="https://2016.igem.org/Team:Oxford/Safety">safety page</a>.</p> | <p>Once the system was able to successfully operate in <i>E. coli</i> DH5-α, we would hope to express it in a strain that more closely mimics the bacteria of the gut. More information on a potential future chassis choice can be found on our <a href="https://2016.igem.org/Team:Oxford/Safety">safety page</a>.</p> | ||
| − | <p>Finally, the design of our system went through many iterations over the summer. One of our most ambitious designs was based on logic gates that would provide an additional safety mechanism | + | <p>Finally, the design of our system went through many iterations over the summer. One of our most ambitious designs was based on logic gates that would provide an additional safety mechanism. We would love to test this system. We discuss this design further on our <a href="https://2016.igem.org/Team:Oxford/Safety">safety page</a>.</p> |
</section> | </section> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/f/f8/Igem2016_532px.jpg" width="50%" /><figcaption>Post presentation photo with the whole team.</figcaption> | ||
<br><br><br> | <br><br><br> | ||
| Line 218: | Line 246: | ||
<li> | <li> | ||
Vita, N., Platsaki, S., Baslé, A., Allen, S.J., Paterson, N.G., Crombie, A.T., Murrell, J.C., Waldron, K.J. and Dennison, C. (2015) ‘A four-helix bundle stores copper for methane oxidation’, Nature, 525(7567), pp. 140–143. doi: 10.1038/nature14854. | Vita, N., Platsaki, S., Baslé, A., Allen, S.J., Paterson, N.G., Crombie, A.T., Murrell, J.C., Waldron, K.J. and Dennison, C. (2015) ‘A four-helix bundle stores copper for methane oxidation’, Nature, 525(7567), pp. 140–143. doi: 10.1038/nature14854. | ||
| + | </li> | ||
| + | <li> | ||
| + | Balogha R., Gyurcsika B., Hunyadi-Gulyásb E., Christensenc H., Jancsóa A., (2016) “Advanced purification strategy for CueR, a cysteine containing copper(I) and DNA binding protein” Protein Expression and Purification Volume 123, July 2016, Pages 90–96. | ||
| + | </li> | ||
| + | <li>Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472.</li> | ||
| + | <li>Philips, S.J., Canalizo-Hernandez M., Yildirim I., Schatz G., Mondragón A., O’Halloran T., (2015) "Allosteric transcriptional regulation via changes in the overall topology of the core promoter." Science 349: 877-881</li> | ||
| + | <li> | ||
| + | http://bionumbers.hms.harvard.edu/KeyNumbers.aspx | ||
| + | </li> | ||
| + | <li> | ||
| + | Selinger, D et al. (2003) Global RNA Half-Life Analysis in Escherichia coli Reveals Positional Patterns of Transcript Degradation, Genome Res. 13, pp. 216-223. | ||
| + | </li> | ||
| + | <li> | ||
| + | http://homepages.ulb.ac.be/~dgonze/BIONUMBERS/bionumbers.html | ||
| + | </li> | ||
| + | <li> | ||
| + | Changela, A et al. (2003) Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR, Science 301 (5638), pp. 1383-1387. | ||
| + | </li> | ||
| + | <li> | ||
| + | Stoyanov, J et al. (2001) CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA, Molecular Microbiology 39(2), pp. 502-512. | ||
| + | </li> | ||
| + | <li> | ||
| + | Gudipaty, S et al (2014) The histidine kinase CusS senses silver ions through direct binding by its sensor domain, Biochim. Biophys. Acta 1844(9), pp. 1656-1661. | ||
| + | </li> | ||
| + | <li> | ||
| + | Hsiao, V et al. (2013) Design and implementation of a synthetic biomolecular concentration tracker, ACS Synth. Biol. 4(2), pp. 150-161. | ||
| + | </li><li> | ||
| + | Wang, B et al. (2013) A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals, Biosensors and Bioelectronics 40, pp. 368-376. | ||
| + | </li> | ||
| + | <li> | ||
| + | Vita, N et al. (2015) A four-helix bundle stores copper for methane oxidation, Nature 525, pp. 140-143. | ||
| + | </li> | ||
| + | <li> | ||
| + | http://physiologyplus.com/motility-in-the-small-intestine/ | ||
| + | </li> | ||
| + | <li> | ||
| + | Moxon, T et al. (2016) In silico modelling of mass transfer & absorption in the human gut, Journal of Food Engineering 176, pp. 110-120. | ||
| + | </li> | ||
| + | <li> | ||
| + | http://cds.ismrm.org/protected/10MProceedings/files/158_7021.pdf flow speed | ||
| + | </li> | ||
| + | <li> | ||
| + | http://trove.nla.gov.au/work/11977899?q&versionId=45681698 other dimensions of gut | ||
</li> | </li> | ||
</ul> | </ul> | ||
Latest revision as of 23:14, 16 November 2016
We were nominated for: Best Therapeutic Project,
Best Wiki, Best Presentation and
Best Education & Public Engagement!
Overview
Rare, or “orphan”, diseases are frequently ignored by the pharmaceutical industry. They encompass a huge range of disorders from ALS to Tourette’s Syndrome, but individually have a relatively low number of patients. The low patient numbers mean that there is very little impetus for the pharmaceutical industry to research and produce novel, innovative therapeutics. This means that patients are often left with unsatisfactory treatments. Our goal is to produce a probiotic therapeutic to treat one such disorder: Wilson’s Disease.
Wilson's Disease
Wilson’s Disease is a genetic disorder characterised by an inability of the body to metabolise copper. Normally, when copper is ingested, it is taken up by the small intestine, taken to the liver and subsequently transported into the blood or excreted into the bile. In Wilson’s Disease, there is a mutation in the gene ATP7B. ATP7B encodes a copper-transporting protein that is responsible for loading copper onto caeruloplasmin for transport in the blood, and into the bile for removal from the body. In the absence of a functional form of this protein, copper is unable to be removed from the liver after absorption. This results in toxic accumulation, as cuprous ions react with hydrogen peroxide to produce dangerous free radicals that damage tissue. This allows copper ions to leak into the blood and eventually accumulate in, and damage, other tissues, such as the kidneys and brain. You can read more about Wilson's disease here.
Current treatments are regarded by patients as unsatisfactory. From discussions with patients, we have identified three main problems with current treatments:
Our Cure aims to address these limitations.

Probiotics
A probiotic is a microorganism that is introduced into the body for its beneficial properties. The concept of a probiotic, meaning “for life”, was introduced by Elie Metchnikoff in 1907, when he hypothesised that replacing or diminishing the populations of ‘putrefactive’ bacteria in the gut with lactic acid bacteria could positively affect bowel health.
Products that are commonly sold as probiotics include foodstuffs, such as yoghurts and cheeses. However, recently there has been an increase in the amount of research going into the use of probiotics as therapeutics, with the genetic engineering of organisms to produce useful substances. Currently there is limited legislation regarding probiotics, as probiotics sold as dietary supplements do not require FDA approval. A genetically-engineered probiotic therapeutic would require more stringent legislation and FDA approval to ensure, through clinical trials, that it works as expected.
Although advancing rapidly, the field of probiotics still requires significant research particularly in areas such as safety. Although regarded as safe for relatively healthy humans to consume, there have been some reports of probiotic-related side effects in people with serious underlying medical conditions. We carried out a comprehensive safety review when completing our project.
Video
Parts
In order to provide a long-term treatment for Wilson's disease we realised that we needed to design components of a bacterial system that could detect copper and produce a copper chelator to prevent its absorption by the body.
We investigated copper detection using promoter systems based upon the native E. coli copper regulator CueR and the CusS/CusR two component system. The two copper chelators we tested were Copper storage protein 1 (Csp1) from Methylosinus trichosporium OB3b and mycobacterial metallothionein (MymT) from Mycobacterium tuberculosis.
You can read about how we chose our parts here.
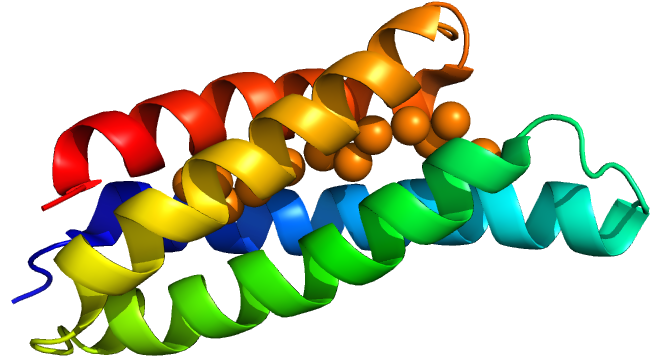
Delivery
From discussion with patients and the public, and the work carried out by previous Oxford iGEM teams, we decided to investigate the use of a bead to deliver our bacteria to the small intestine. Our bacteria will initially be encapsulated in an alginate matrix, and then be alternately coated in layers of alginate and chitosan. The goal of the multiple polymer-coatings is to protect the bacteria from the harsh conditions of the stomach, whilst having the ability to degrade in the more alkaline pH of the small intestine. This degradation releases our bacteria into the favourable conditions of the small intestine, where they can colonise the area and chelate dietary copper.

Results
Through our experimental work we have been able to obtain data suggesting the following:
- Different arrangements of both our CueR-linked and CusS/CusR-linked promoter systems are sensitive over a range of copper concentrations, including at the lower concentrations mimicking the gut.
- MymT is able to chelate a measurable amount of copper in vivo.
- Alternately layered chitosan-alginate beads release material in the small intestine, following passage through the stomach, in a simulation accounting for the pH change.
Please see our experiments and results pages for more discussion of our methods and the outcomes.
Improving Registry Parts
As part of our project, we have improved the function/characterisation of three poorly documented parts.
Please visit our parts and results pages for further information.
BBa_I760005:
This copy of pCusC located in the registry had been previously used 11 times, but had no associated useful characterisation data. From discussions with a mentor, Tom Folliard, we came to the conclusion that this may have been because the promoter region was too short, and thus missing key binding sites. We decided to deposit an elongated form of this promoter, which we have achieved thorough characterisation.

BBa_K190020:
MymT can be found in the registry with no associated characterisation data. We codon-optimised the sequence for expression in E. coli and added a hexahistidine tag for purification. We also produced a form with a C-terminal sfGFP tag (also his-tagged) to aid purification, allowing us to view the cellular distribution of MymT and allow analysis by Fluorescence Lifetime IMaging (FLIM). The fluorescence lifetime of GFP with a C-terminal His tag was shown to be reduced by copper by Hötzer et al., allowing FLIM to be used in an in vivo assay of copper binding. This has provided us with preliminary characterisation data for the protein, suggesting copper-binding activity in vivo. More details can be found on our results page.

BBa_K1758324:
We were interested in using this part from Team Bielefeld-CeBiTec in 2015 as a potential copper biosensor.
The team assembled a copper biosensor from two subparts which they then joined together:
The first part is a pCopA-RBS-sfGFP and the second part is the regulator CueR expressed from a constitutive promoter. The part is deposited in the registry and labelled to suggest the CueR is expressed divergent from the sfGFP (on the opposite strand and transcribed in the opposite direction):

However if you look at the sequence level this is clearly not the case. The constitutive promoter and the CueR start codon are at the 5’ end of the sfGFP coding strand and the CueR stop codon just upstream of pCopA. The part in fact has the constitutive promoter on the same strand as pCopA and sfGFP facing in the same direction and would be better represented like this:

As the two coding regions are not separated by a transcription terminator, there would be read through from the constitutive promoter to the sfGFP and sfGFP would be expressed even in the absence of copper. As no negative control is included in the plate reader graph they provide and no settings provided for their BioLector experiments in their protocols it is unclear just how high the expression level at 0mM copper was for this part compared to a negative control strain.
The CueR subpart (BBa_K1758320) making up BBa_K1758324 is also incorrectly labelled.
When we designed this part we flipped the CueR and the constitutive promoter to face the opposite direction on the opposite strand i.e. so they were divergent. We also had to remove the 5'UTR, which Bielefeld found to increase expression, because it was too AT rich to be synthesised.
Unfortunately, every attempt to amplify this part from the synthesised sequence we received from IDT resulted in the same two point mutations in the sfGFP region of this part making it non-functional. To compare this promoter system to the others we designed, we used our parts with chelator-sfGFP fusions instead of the sfGFP which we expected to have similar behaviour.
Conclusion
Over the course of the summer we have successfully created and submitted 13 sequence-confirmed BioBrick parts, 8 of which have been characterised. Through our experiments, we have been able to examine the copper-sensitivity of a variety of promoters integral to our Cure. In addition, we have tested the chelating-ability of our copper chelators with a variety of different assays. Although these have not all been successful, through a collaboration with Cardiff we have been able to obtain preliminary data suggesting that MymT may be able to successfully lower the intracellular copper concentration. We have also obtained promising preliminary data relating to the delivery of substances to the small intestine via alginate-chitosan beads.
In conclusion, over the course of the summer we have successfully managed to produce an initial proof of concept for our idea and have contributed to our understanding of these systems, in addition to investigating a treatment that addresses the needs and concerns of the patients themselves.
Future
To further develop our project, we would hope to carry out a number of further experiments to allow us to generate something more than a proof-of-concept model. Primarily, we would hope to properly characterise the copper-chelating ability of Csp1 and confirm its localisation in the periplasm by redesigning the TAT sequence. Additionally, we would carry out further experiments to refine our delivery system.
Once the system was able to successfully operate in E. coli DH5-α, we would hope to express it in a strain that more closely mimics the bacteria of the gut. More information on a potential future chassis choice can be found on our safety page.
Finally, the design of our system went through many iterations over the summer. One of our most ambitious designs was based on logic gates that would provide an additional safety mechanism. We would love to test this system. We discuss this design further on our safety page.

References:
- Roberts, E. (2011) ‘Wilson’s Disease’, Medicine, 39(10), pp. 602–604.
- Schilsky, M.L., Roberts, E.A., Hahn, S. and Askari, F. (2015) ‘Costly choices for treating Wilson’s disease’, Hepatology, 61(4), pp. 1106–1108. doi: 10.1002/hep.27663.
- Czlonkowska, A., Gajda, J. and Rodo, M. (1996) ‘Effects of long-term treatment in Wilson’s disease with d-penicillamine and zinc sulphate’, Journal of Neurology, 243(3), pp. 269–273. doi: 10.1007/bf00868525.
- Duan, F.F., Liu, J.H. and March, J.C. (2015) ‘Engineered Commensal bacteria Reprogram intestinal cells into glucose-responsive Insulin-Secreting cells for the treatment of diabetes’, Diabetes, 64(5), pp. 1794–1803. doi: 10.2337/db14-0635.
- Didari, T., Solki, S., Mozaffari, S., Nikfar, S. and Abdollahi, M. (2014) ‘A systematic review of the safety of probiotics’, Expert Opinion on Drug Safety, 13(2), pp. 227–239. doi: 10.1517/14740338.2014.872627.
- Anukam K. C., Reid G. (2007). “Probiotics: 100 years (1907–2007) after Elie Metchnikoff’s Observation,” Communicating Current Research and Educational Topics and Trends in Applied Microbiology, ed. Méndez-Vilas A., editor. (Formatex.org) 466–474
- Cook, M.T., Tzortzis, G., Khutoryanskiy, V.V. and Charalampopoulos, D. (2013) ‘Layer-by-layer coating of alginate matrices with chitosan–alginate for the improved survival and targeted delivery of probiotic bacteria after oral administration’, J. Mater. Chem. B, 1(1), pp. 52–60. doi: 10.1039/c2tb00126h.
- Chávarri, M., Marañón, I., Ares, R., Ibáñez, F.C., Marzo, F. and Villarán, M. del C. (2010) ‘Microencapsulation of a probiotic and prebiotic in alginate-chitosan capsules improves survival in simulated gastro-intestinal conditions’, International Journal of Food Microbiology, 142(1-2), pp. 185–189. doi: 10.1016/j.ijfoodmicro.2010.06.022.
- Krasaekoopt, W., Bhandari, B. and Deeth, H. (2004) ‘The influence of coating materials on some properties of alginate beads and survivability of microencapsulated probiotic bacteria’,International Dairy Journal, 14(8), pp. 737–743. doi: 10.1016/j.idairyj.2004.01.004.
- Argüello, J.M., Raimunda, D. and Padilla-Benavides, T. (2013) ‘Mechanisms of copper homeostasis in bacteria’, Frontiers in Cellular and Infection Microbiology, 3. doi: 10.3389/fcimb.2013.00073.
- Grey, B. and Steck, T.R. (2001) ‘Concentrations of copper thought to be toxic to Escherichia coli can induce the viable but Nonculturable condition’, Applied and Environmental Microbiology, 67(11), pp. 5325–5327. doi: 10.1128/aem.67.11.5325-5327.2001.
- Lewis, K.O. (1973) ‘The nature of the copper complexes in bile and their relationship to the absorption and excretion of copper in normal subjects and in Wilson’s disease’, Gut, 14(3), pp. 221–232. doi: 10.1136/gut.14.3.221.
- Rensing, C. and Grass, G. (2003) ‘Escherichia coli mechanisms of copper homeostasis in a changing environment’, FEMS Microbiology Reviews, 27(2-3), pp. 197–213. doi: 10.1016/s0168-6445(03)00049-4.
- Zoetendal, E.G., Raes, J., van den Bogert, B., Arumugam, M., Booijink, C.C., Troost, F.J., Bork, P., Wels, M., de Vos, W.M. and Kleerebezem, M. (2012) ‘The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates’, The ISME Journal, 6(7), pp. 1415–1426. doi: 10.1038/ismej.2011.212.
- Franz, K.J. (2012) ‘Application of inorganic chemistry for non-cancer therapeutics’, Dalton Transactions, 41(21), p. 6333. doi: 10.1039/c2dt90061k.
- Das, S.K. and Ray, K. (2006) ‘Wilson’s disease: An update’, Nature Clinical Practice Neurology, 2(9), pp. 482–493. doi: 10.1038/ncpneuro0291.
- Yamamoto, K. and Ishihama, A. (2005) ‘Transcriptional response of Escherichia coli to external copper’, Molecular Microbiology, 56(1), pp. 215–227. doi: 10.1111/j.1365-2958.2005.04532.x.
- Gold, B., Deng, H., Bryk, R., Vargas, D., Eliezer, D., Roberts, J., Jiang, X. and Nathan, C. (2008) ‘Identification of a copper-binding metallothionein in pathogenic mycobacteria’, Nature chemical biology, 4(10), pp. 609–616. doi: 10.1038/nchembio.109.
- Outten, F.W. (2001) ‘The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli’, Journal of Biological Chemistry, 276(33), pp. 30670–30677. doi: 10.1074/jbc.m104122200.
- Ravikumar, S., Pham, V.D., Lee, S.H., Yoo, I. and Hong, S.H. (2012) ‘Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop’, Journal of Industrial Microbiology & Biotechnology, 39(6), pp. 861–868. doi: 10.1007/s10295-012-1096-y.
- Neupert, J., Karcher, D. and Bock, R. (2008) ‘Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli’, Nucleic Acids Research, 36(19), pp. e124–e124. doi: 10.1093/nar/gkn545.
- Vita, N., Platsaki, S., Baslé, A., Allen, S.J., Paterson, N.G., Crombie, A.T., Murrell, J.C., Waldron, K.J. and Dennison, C. (2015) ‘A four-helix bundle stores copper for methane oxidation’, Nature, 525(7567), pp. 140–143. doi: 10.1038/nature14854.
- Balogha R., Gyurcsika B., Hunyadi-Gulyásb E., Christensenc H., Jancsóa A., (2016) “Advanced purification strategy for CueR, a cysteine containing copper(I) and DNA binding protein” Protein Expression and Purification Volume 123, July 2016, Pages 90–96.
- Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472.
- Philips, S.J., Canalizo-Hernandez M., Yildirim I., Schatz G., Mondragón A., O’Halloran T., (2015) "Allosteric transcriptional regulation via changes in the overall topology of the core promoter." Science 349: 877-881
- http://bionumbers.hms.harvard.edu/KeyNumbers.aspx
- Selinger, D et al. (2003) Global RNA Half-Life Analysis in Escherichia coli Reveals Positional Patterns of Transcript Degradation, Genome Res. 13, pp. 216-223.
- http://homepages.ulb.ac.be/~dgonze/BIONUMBERS/bionumbers.html
- Changela, A et al. (2003) Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR, Science 301 (5638), pp. 1383-1387.
- Stoyanov, J et al. (2001) CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA, Molecular Microbiology 39(2), pp. 502-512.
- Gudipaty, S et al (2014) The histidine kinase CusS senses silver ions through direct binding by its sensor domain, Biochim. Biophys. Acta 1844(9), pp. 1656-1661.
- Hsiao, V et al. (2013) Design and implementation of a synthetic biomolecular concentration tracker, ACS Synth. Biol. 4(2), pp. 150-161.
- Wang, B et al. (2013) A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals, Biosensors and Bioelectronics 40, pp. 368-376.
- Vita, N et al. (2015) A four-helix bundle stores copper for methane oxidation, Nature 525, pp. 140-143.
- http://physiologyplus.com/motility-in-the-small-intestine/
- Moxon, T et al. (2016) In silico modelling of mass transfer & absorption in the human gut, Journal of Food Engineering 176, pp. 110-120.
- http://cds.ismrm.org/protected/10MProceedings/files/158_7021.pdf flow speed
- http://trove.nla.gov.au/work/11977899?q&versionId=45681698 other dimensions of gut

