AlexAlario (Talk | contribs) |
AlexAlario (Talk | contribs) |
||
| Line 97: | Line 97: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/0/00/T--Austin_UTexas--IsolatingStrains.png" style="width:700px;"> | <img src="https://static.igem.org/mediawiki/2016/0/00/T--Austin_UTexas--IsolatingStrains.png" style="width:700px;"> | ||
| − | <figcaption><b>Figure 1:</b> Shows YPD plates spread with various dilutions of GT's brand kombucha samples. Credit: Zach Martinez</figcaption> | + | <figcaption><b>Figure 1:</b> Shows YPD plates spread with various dilutions of GT's brand kombucha samples. Credit: Zach Martinez.</figcaption> |
</figure> | </figure> | ||
</center> | </center> | ||
| Line 203: | Line 203: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/3/3f/T--Austin_UTexas--MIC.png" style="width:800px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/3/3f/T--Austin_UTexas--MIC.png" style="width:800px;display:inline-block"> | ||
| − | <figcaption><b>Figure 2:</b> These are a Minimum Inhibitory Concentration experiment that included using kanamycin, spectinomycin and carbenicillin with <i>G. oxydans</i> in order to observe if the strain can resist standard concentrations of antibiotics used with <i>E. coli</i>. The top row of cultures are <i>G. oxydans</i> with concentrations of carbenicillin ranging from 4x to none (1x = 100µg/mL). This set of tubes show that <i>G. oxydans</i> is resistant to carbenicillin, at least up to a 4x concentration. The next row is <i>G. oxydans</i> in concentrations of kanamycin from 4x to none (1x = 50µg/mL). This set of reactions demonstrates how <i>G. oxydans</i> is resistant to kanamycin, up to a 1x concentration. The last row of tubes is <i>G. oxydans</i> in concentrations of spectinomycin ranging from 4x to none (1x = 60µg/mL). This last row shows how <i>G. oxydans</i> is resistant to spectinomycin, at least up to a 4x concentration. Credit: Zach Martinez </figcaption> | + | <figcaption><b>Figure 2:</b> These are a Minimum Inhibitory Concentration experiment that included using kanamycin, spectinomycin and carbenicillin with <i>G. oxydans</i> in order to observe if the strain can resist standard concentrations of antibiotics used with <i>E. coli</i>. The top row of cultures are <i>G. oxydans</i> with concentrations of carbenicillin ranging from 4x to none (1x = 100µg/mL). This set of tubes show that <i>G. oxydans</i> is resistant to carbenicillin, at least up to a 4x concentration. The next row is <i>G. oxydans</i> in concentrations of kanamycin from 4x to none (1x = 50µg/mL). This set of reactions demonstrates how <i>G. oxydans</i> is resistant to kanamycin, up to a 1x concentration. The last row of tubes is <i>G. oxydans</i> in concentrations of spectinomycin ranging from 4x to none (1x = 60µg/mL). This last row shows how <i>G. oxydans</i> is resistant to spectinomycin, at least up to a 4x concentration. Credit: Zach Martinez. </figcaption> |
</figure> | </figure> | ||
</center> | </center> | ||
| Line 228: | Line 228: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/3/38/T--Austin_UTexas--PreviousRecaps.png" style="width:600px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/3/38/T--Austin_UTexas--PreviousRecaps.png" style="width:600px;display:inline-block"> | ||
| − | <figcaption><b>Figure 1:</b> Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls. Credit: Katelyn Corley</figcaption> | + | <figcaption><b>Figure 1:</b> Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls. Credit: Katelyn Corley.</figcaption> |
</figure> | </figure> | ||
<br><br> | <br><br> | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/d/d6/T--Austin_UTexas--OngoingRecapitulations.png" style="width:900px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/d/d6/T--Austin_UTexas--OngoingRecapitulations.png" style="width:900px;display:inline-block"> | ||
| − | <figcaption><b>Figure 2:</b> Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 7 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after 4 days, forming a mature pellicle by Day 7. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated both microbes that had been purchased and microbe that had been isolated from kombucha itself. Row 3 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is thick and has multiple carbon dioxide bubbles. Credit: Katelyn Corley</figcaption> | + | <figcaption><b>Figure 2:</b> Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 7 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after 4 days, forming a mature pellicle by Day 7. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated both microbes that had been purchased and microbe that had been isolated from kombucha itself. Row 3 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is thick and has multiple carbon dioxide bubbles. Credit: Katelyn Corley.</figcaption> |
</figure> | </figure> | ||
</center> | </center> | ||
| Line 261: | Line 261: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/2/24/T-Austin_UTexas--File_ADHblasttable.png" style="width:500px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/2/24/T-Austin_UTexas--File_ADHblasttable.png" style="width:500px;display:inline-block"> | ||
| − | <figcaption><b>Table 1:</b> Results of BLAST search comparing the amino acid sequence for PQQ-ADH in C. testosteroni against similar amino acid sequences in Komagataeibacter xylinus (identical to Ga. hansenii). Line 3 is a close match, and the accession number matches one of the ADH genes found in K. xylinus.</figcaption> | + | <figcaption><b>Table 1:</b> Results of BLAST search comparing the amino acid sequence for PQQ-ADH in C. testosteroni against similar amino acid sequences in Komagataeibacter xylinus (identical to Ga. hansenii). Line 3 is a close match, and the accession number matches one of the ADH genes found in K. xylinus. Credit: Stratton Georgoulis.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 276: | Line 276: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/6/69/T--Austin_UTexas--ALDHlinmap.png" style="width:600px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/6/69/T--Austin_UTexas--ALDHlinmap.png" style="width:600px;display:inline-block"> | ||
| − | <figcaption><b>Figure 2:</b> Linear map of the coding sequence for membrane-bound ALDH with a Golden Gate type 3 prefix and suffix. BsmBI and BsaI sites are indicated. The restriction sites at either end are included in the prefix and suffix, but the internal BsaI site must be removed to create a functioning Golden Gate part.</figcaption> | + | <figcaption><b>Figure 2:</b> Linear map of the coding sequence for membrane-bound ALDH with a Golden Gate type 3 prefix and suffix. BsmBI and BsaI sites are indicated. The restriction sites at either end are included in the prefix and suffix, but the internal BsaI site must be removed to create a functioning Golden Gate part. Credit: Stratton Georgoulis.</figcaption> |
</figure> | </figure> | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/d/dc/T--Austin_UTexas--ADHlinmap.png" style="width:600px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/d/dc/T--Austin_UTexas--ADHlinmap.png" style="width:600px;display:inline-block"> | ||
| − | <figcaption><b>Figure 3:</b> Linear map of the coding sequence for PQQ-ADH with a Golden Gate type 3 prefix and suffix. EcoRI, BsmBI, and BsaI sites are indicated. The centermost BsmBI restriction site is in the coding sequence and must be removed to create a functional Golden Gate part. EcoRI is not used in Golden Gate assembly, so those sites do not necessarily need to be removed.</figcaption> | + | <figcaption><b>Figure 3:</b> Linear map of the coding sequence for PQQ-ADH with a Golden Gate type 3 prefix and suffix. EcoRI, BsmBI, and BsaI sites are indicated. The centermost BsmBI restriction site is in the coding sequence and must be removed to create a functional Golden Gate part. EcoRI is not used in Golden Gate assembly, so those sites do not necessarily need to be removed. Credit: Stratton Georgoulis.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 289: | Line 289: | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2016/a/a9/T--Austin_UTexas--oligotable.png" style="width:600px;display:inline-block"> | <img src="https://static.igem.org/mediawiki/2016/a/a9/T--Austin_UTexas--oligotable.png" style="width:600px;display:inline-block"> | ||
| − | <figcaption><b>Table 2:</b> Description of oligonucleotides ordered from IDT and their purposes. All of these are PCR primers except for igem2016_KOM_EtOH_07, which is a gBlock containing the end of the PQQ-ADH with a Golden Gate type 3 suffix appended.</figcaption> | + | <figcaption><b>Table 2:</b> Description of oligonucleotides ordered from IDT and their purposes. All of these are PCR primers except for igem2016_KOM_EtOH_07, which is a gBlock containing the end of the PQQ-ADH with a Golden Gate type 3 suffix appended. Credit: Stratton Georgoulis.</figcaption> |
</figure> | </figure> | ||
</center> | </center> | ||
Revision as of 23:02, 19 October 2016
Results

Click on one of the images below to learn more about our results!
GOX Sequences as Putative Promoters
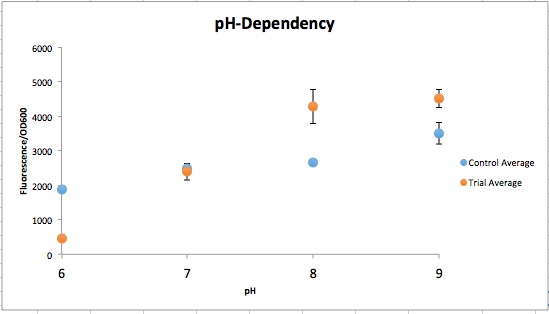
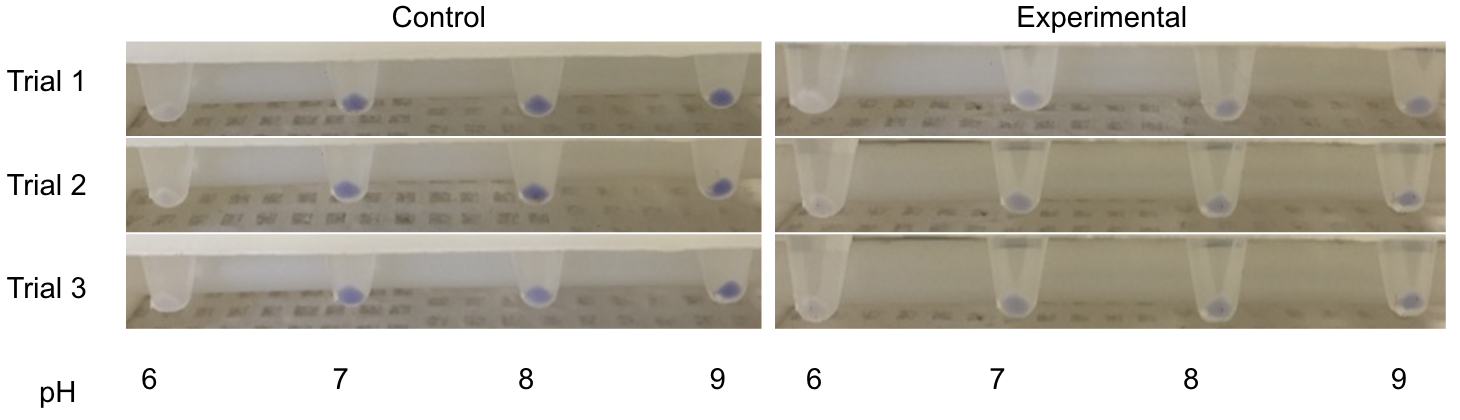
Three endogenous upstream regions of loci on the Gluconobacter oxydans chromosome were reported to show increased mRNA synthesis as pH decreased, were isolated and obtained, as seen in table 1 (Hanke, et al., 2012). Using Golden Gate assembly, these putative promoters have been placed on the Golden Gate entry vector pYTK001 for later use. By utilizing these pH-sensitive promoters with different reporters and transforming them into multiple organisms in kombucha, the visualization of the microbes and their location in kombucha would be possible (Lee, et al., 2015). This will serve as a stepping stone into further understanding how the microbiome of kombucha changes as it brews as well as determining organism concentration specific times during the brewing process.
| Locus Tag | Predicted Functions | mRNA ratio pH4/pH6 |
|---|---|---|
| GOX0647 | Putative exporter protein, ArAE family | 12.91 |
| GOX0890 | Hypothetical protein GOX0890 | 4.93 |
| GOX1841 | Hypothetical protein GOX1841 | 3.36 |
References
- The Barrick Lab Conjugation Protocol
- Abbot, J. Komagataeibacter xylinus isolate ATCC53582 genome assembly, contig: ATCC53582_Chromosome, whole genome shotgun sequence. 2015. Accessed from NCBI website.
- BIT-China-2015
- Calloway, Ewen. (2015) Lab staple agar hit by seaweed shortage. Nature.
- Hanke, T., Richhardt, J., Polen, T., Sahm, H., Bringer, S., and Bott, M. (2012) Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. Journal of Biotechnology 157, 359–372.
- Ioannis Giavasis et al. (2000) Gellan Gum Critical Reviews in Biotechnology., 20.3: 177-211
- Kang, Kenneth S. et al. (1982) Agar-Like Polysaccharide Produced by a Pseudomonas Species: Production and Basic Properties. Applied and Environmental Microbiology., 1086-1091
- Kuper, C., and Jung, K. (2005) CadC-mediated activation of the cadBA promoter in Escherichia coli. Journal of Molecular and Microbiological Biotechnology 1, 26–39.
- Lee ME, DeLoache, WC A, Cervantes B, Dueber, JE. (2015) A Highly-characterized Yeast Toolkit for Modular, Multi-part Assembly. ACS Synthetic Biology 4 975-986
- Mamlouk, Y. and M. Gullo. Acetic Acid Bacteria: Physiology and carbon sources oxidation. 2013. Indian Journal of Microbiology 53 (4): 337-384.
- Nakayama, S.-I., and Watanabe, H. (1998) Identification of cpxR as a Positive Regulator Essential for Expression of the Shigella sonnei virF Gene. Journal of Bacteriology 180, 3522–3528.
- Nakayama, S.-I., and Watanabe, H. (1995) Involvement of cpxA, a Sensor of a Two-Component Regulatory System, in the pH-Dependent Regulation of Expression of Shigella sonnei virF Gene. Journal of Bacteriology 177, 5062–5069.
- Robillard, R. A microbial breathalyzer: design of a colorimetric assay for the detection and quantification of ethanol production in microbes. 2007. Major qualifying project for a B.S. degree from Worcester Polytechnic Institute.
- Wang, Xia, et al. (2006) Modeling for Gellan Gum Production by Sphingomonas paucimobilis ATCC 31461 in a Simplified Medium. Applied and Environmental Microbiology, 3367-3374
- Wu et. al. (2014) Yellow pigments generation deficient Sphingomonas strain and application thereof in Gellan Gum. US Patent 8,685,698.
Back to Top