Introduction
Last year we introduced the concept of Flavorator.
Here, we explain Flavorator's concept.
Food problems are serious matters in the world.
Among them is food preservation.
Ideal food preservation is keeping food without causing quality change for a long time and at low cost.
Various preservation methods have been used so far, and we created a new one.
It is "Flavorator"!!
Thirty years ago, “KOZOKO(香蔵庫 in Japanese)” was proposed by Professor Yozo Iwanami.
The name “KOZOKO” can be directly translated into ‘flavor (=KO, 香)’- ‘preserved (=ZO, 蔵)’- ‘box (=KO, box)’. However, “KOZOKO” is not just a flavor-keeping box but a box to preserve food in a fragrance.
Then, what flavors should we put in KOZOKO?
The flavors should have antimicrobial or insecticidal activities.
The best candidates are those of plant origin.
Plants cannot move to other places even when microbes or insects attack them. So, they produce antimicrobial or insecticidal agents.
These agents are mostly not harmful to humans and animals.
Therefore, “KOZOKO” with antimicrobial flavors would be an energy-saving substitute for an ordinary electric refrigerator.
In order to make “KOZOKO” practical two problems have to be solved.
First, we must select appropriate flavors and plants that can be cultivated under the various climate conditions.
Second, cost issues must be cleared.
Mass cultivation of plants and extraction of pure flavors from plants are time-consuming and expensive.
Consequently, at present “KOZOKO” is under the state of conceptual idea.
We created a new version of “KOZOKO” utilizing syntheticbiology.
We named it “Flavorator”.
And we want to solve Food problem all over the world!
Recombinant Escherichia coli produces antimicrobial and insecticidal volatile gaseous substances that suppress bacterial growth in the “Flavorator” and prevent insects from entering the “Flavorator”.
In the future, “Flavorator” can slow down food decay and solve one of the food problems. This year, we started an action towards improvement of Flavorator.
We use E. coli as a model organism to show feasibility of Flavorator.
We decided to do the following.
First, we use high antimicrobial fragrance to synthesis easily in E. coli.
Second, we need to optimize E.coli to Flavorator.
○First, we chose farnesol as an effective fragrance which can be synthesized easily by E.coli. .
There are three reasons for this choice.
1. We can increase production of farnesol by overexpression of E.coli's endogenous gene because E. coli can synthesize farnesol in theory.
2. Farnesol does not impair flavor of food because it is almost odorless.
3. Farnesol has a high antibacterial activity against food poisoning bacteria.
◯Second, we did three designs ,upon optimization of E.coli.
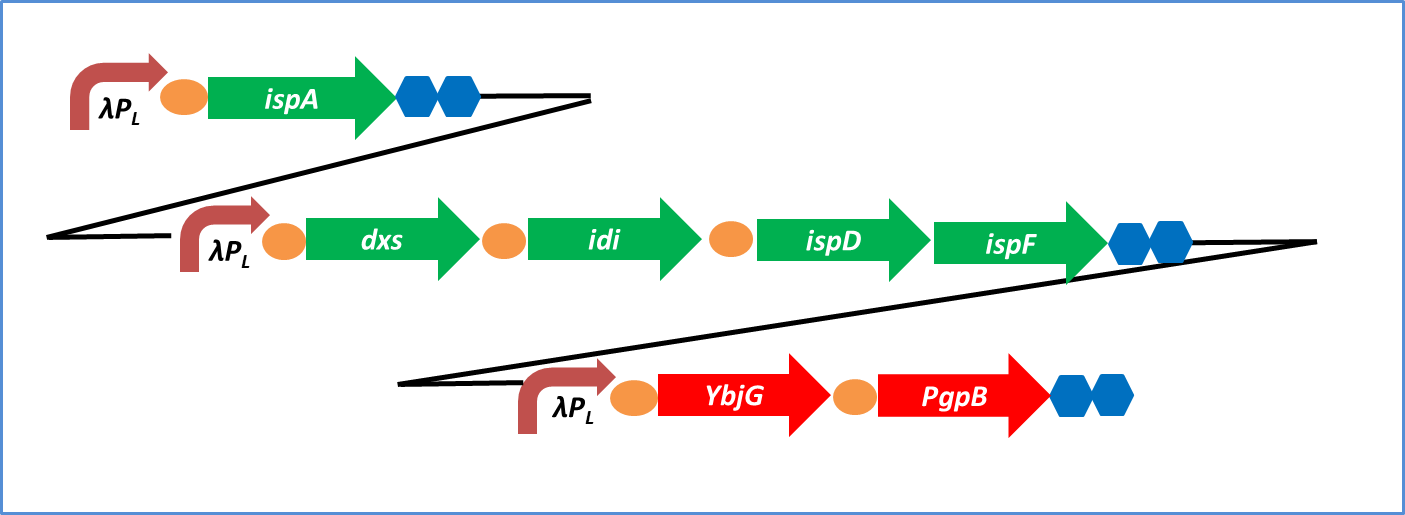
1. Construction of farnesol synthetic plasmid for E. coli
Endogenous phosphatase of Escherichia coli is not able to synthesize much farnesol.
Therefore, we introduced additional endogenous farnesol synthesis enzyme genes which are PgpB and YbjG in E.coli.
We added PgpB and YbjG to device of the last year
Ybjg and PgpB encode farnesol synthase.
Farnesol synthase dephosphorylates farnesyl diphosphate.
As a result, this device can synthesize farnesol.
2. Improved resistance of E. coli to farnesol
We further introduce an activator gene of AcrAB-TolC efflux pump (MarA) to We further introduce an activator gene of AcrAB-TolC efflux pump (MarA). This improves the resistance of E. coli against farnesol.
3. Addition of metabolic pathway from xylose genes.
The m-ribB (1-Deoxy-D-xylulose 5-phosphate synthase gene) which is mutant of ribB mutated G108S 1-Deoxy-D-xylulose 5-phosphate synthase can utilize D-ribulose 5-phosphate as substrates, generating 1-Deoxy-D-xylulose 5-phosphate.
Ybjg and PgpB encode farnesol synthase.
Farnesol synthase dephosphorylates farnesyl diphosphate.
As a result, this device can synthesize farnesol.
The three designs above are methods to transform plasmids into E. coli.
In other words, we use these as recombinants to "Flavorator".
We are forbidden to put E.coli outside of the laboratory.
It is difficult to realize "Flavorator" the way things are going.
So we used the CRISPR-Cas9 to alter the genome of E. coli.
Accordingly, we thought preventing recombinant leaking to the outside of the laboratory.
For example, we thought about trying to knock out the genes involved in the growth of E. coli, so that E. coli cannot grow except for a particular medium.
We tried to realize "Flavorator" in the above-described method.
Therefore, we do knockout of an endogenous gene of E. coli using the CRISPR-Cas9.
By doing this we thought about trying to make more suitable E. coli strain to "Flavorator".
Specifically, we had knocked out unwanted intermediate metabolite synthase gene to synthesize fragrance without incorporating the plasmid.
So we tried to knockout one gene (gdhA) to explain this concept.
Escherichia coli originally synthesizes little farnesol. Therefore, we knocked out intermediate metabolite synthase gene unnecessary for farnesol synthesis in order to facilitate the synthesis proceeding to DXP which is the substrate of farnesol.
CRISPR system is a bacterial immune system which is used as a gene engineer machine now.
It has three types and we chose the type II CRISPR system.
This system contains three parts.
First, this system can express two kinds of single strand RNA: tracrRNA and crRNA.
crRNA’s structure is like “repeat---spacer---repeat”.
The spacer part is complementary to target gene’s DNA sequence, and the repeat is complementary to tracrRNA.
tracrRNA can also have interaction with a protein coded in this system --- Cas9, an endonuclease.
Once we modified the spacer sequence of a CRISPR system and transformed it into bacteria, it will search for target gene Intermediate metabolite synthase gene was not used to synthesize farnesol gene in our project) in bacteria’s genome by crRNA.
Once the target is found, Cas9 protein will bind on this gene’s DNA sequence with the help of tarcrRNA and crRNA.
Then, Cas9 will make a small double strand break on the gene.
So this gene’s sequence will be modified by later homozygous recombination, resulting in the knock-out of this gene.
We have considered trying to be able to use as a strain for Flavorator by changing the genome of E. coli strain.
We showed the possibility to synthesize fragrance without recombinant by using CRISPR-Cas9.
In the future, we will not need to use recombinant E. coli.
REFERENCES
[http://onlinelibrary.wiley.com/doi/10.1002/biot.201600250/abstract?systemMessage=Wiley+Online+Library+will+be+unavailable+on+Saturday+30th+July+2016+from+08:00-11:00+BST+/+03:00-06:00+EST+/+15:00-18:00+SGT+for+essential+maintenance.Apologies+for+the+inconvenience [1] Wang C, Park JE, Choi ES, Kim SW.(2016).Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway – application of PgpB and YbjG phosphatases. Biotechnology Journal 11(10): 1291–1297.
[http://aem.asm.org/content/81/1/130.short [2] Kirby J, Nishimoto M, Chow W. N.R, Baidoo E.E., Wang G, Martin J, … & Keasling J D.(2015).Enhancing Terpene Yield from Sugars via Novel Routes to 1-Deoxy-d-Xylulose 5-Phosphate.Applied and Environmental Microbiology,81(1), 130-138.
[http://link.springer.com/article/10.1007/s00284-009-9408-9 [3]Gomes FIA, Teixeira P, Azeredo J, Oliveira R. (2009). Effect of Farnesol on Planktonic and Biofilm Cells of Staphylococcus epidermidis. Current Microbiology, 59(2), 118-122.
[http://www.nature.com/nbt/journal/v31/n3/full/nbt.2508.html%3FWT.ec_id%3DNBT-201303 [4]Jian W, Bikard D, Cox D, Zhang F & Marraffini LA. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31(3), 233-239.
[http://www.sciencedirect.com/science/article/pii/S1096717615000750 [5]Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang Y, Chen T, Zhao X.(2015). Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metabolic Engineering, 31, 13-21.
[http://www.sciencedirect.com/science/article/pii/S1389172312004185 [6]Shah A A, Wang C, Chung Y R, Kim J Y, Choi E S, Kim S W. (2013).Enhancement of geraniol resistance of Escherichia coli by MarA overexpression. Journal of Bioscience and Bioengineering, 115(3),235–258.
[http://nar.oxfordjournals.org/content/early/2016/04/08/nar.gkw223.abstract [7]Cui L, Bikard D. (2016).Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic acids research, gkw223.
[http://www.sciencedirect.com/science/article/pii/S1096717605000741 [8]Yuan L.Z, Rouvière P.E, LaRossa R.A, Suh W. (2006).Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metabolic Engineering 8(1) 79–90.
[http://aem.asm.org/content/81/15/5103.short [9]Pyne ME, Moo-Young M, Chung DA, Chou CP. 2015. Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Applied and Environmental Microbiology 81:5103–5114.