Stratton g (Talk | contribs) |
|||
| Line 223: | Line 223: | ||
<br> | <br> | ||
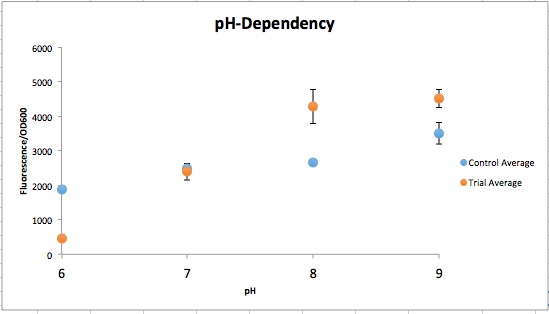
<p>After having the essential kombucha microbes, the next set of recapitulations tested what ratios these microbes needed to be in when placed in tea media so they could properly form a SCOBY and recreate kombucha. This process involved multiple trial and error results, but eventually the results included below were found. | <p>After having the essential kombucha microbes, the next set of recapitulations tested what ratios these microbes needed to be in when placed in tea media so they could properly form a SCOBY and recreate kombucha. This process involved multiple trial and error results, but eventually the results included below were found. | ||
| − | |||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/d/d6/T--Austin_UTexas--OngoingRecapitulations.png" style="width:900px;display:inline-block"> | ||
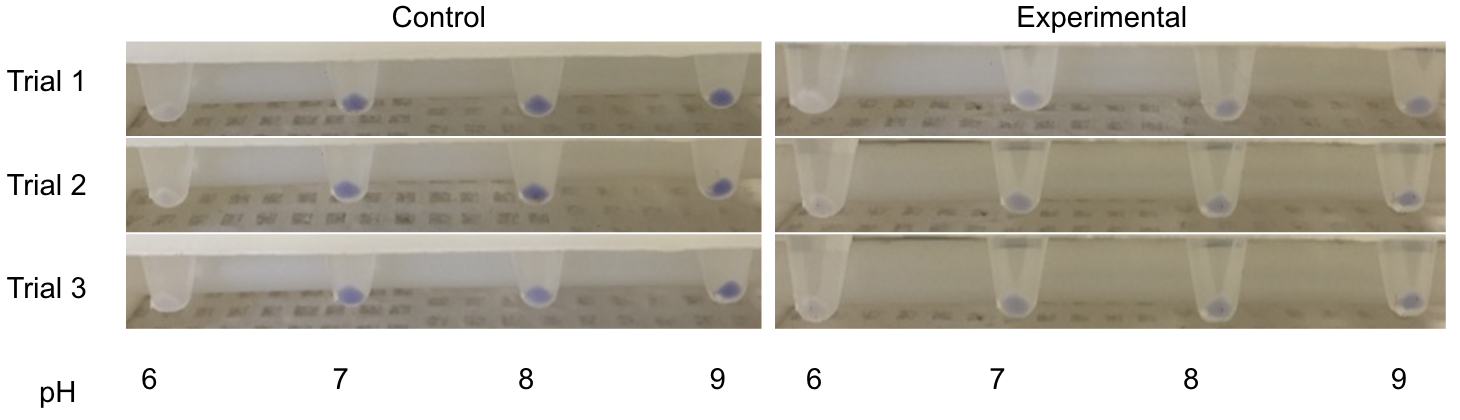
| + | <figcaption><b>Figure 2:</b> Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 7 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after 4 days, forming a mature pellicle by Day 7. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated both microbes that had been purchased and microbe that had been isolated from kombucha itself. Row 3 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is thick and has multiple carbon dioxide bubbles.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </html> | ||
[[File:T--Austin_UTexas--PreviousRecaps.png|thumb|center|640px|Figure 1: Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls.]] | [[File:T--Austin_UTexas--PreviousRecaps.png|thumb|center|640px|Figure 1: Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of <i>Lachancea fermentati</i> each isolated from kombucha samples, as well as a strain of <i>Gluconobacter oxydans</i> and <i>Gluconacetobacter hansenii</i>. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls.]] | ||
| − | |||
| − | |||
<html> | <html> | ||
| Line 255: | Line 258: | ||
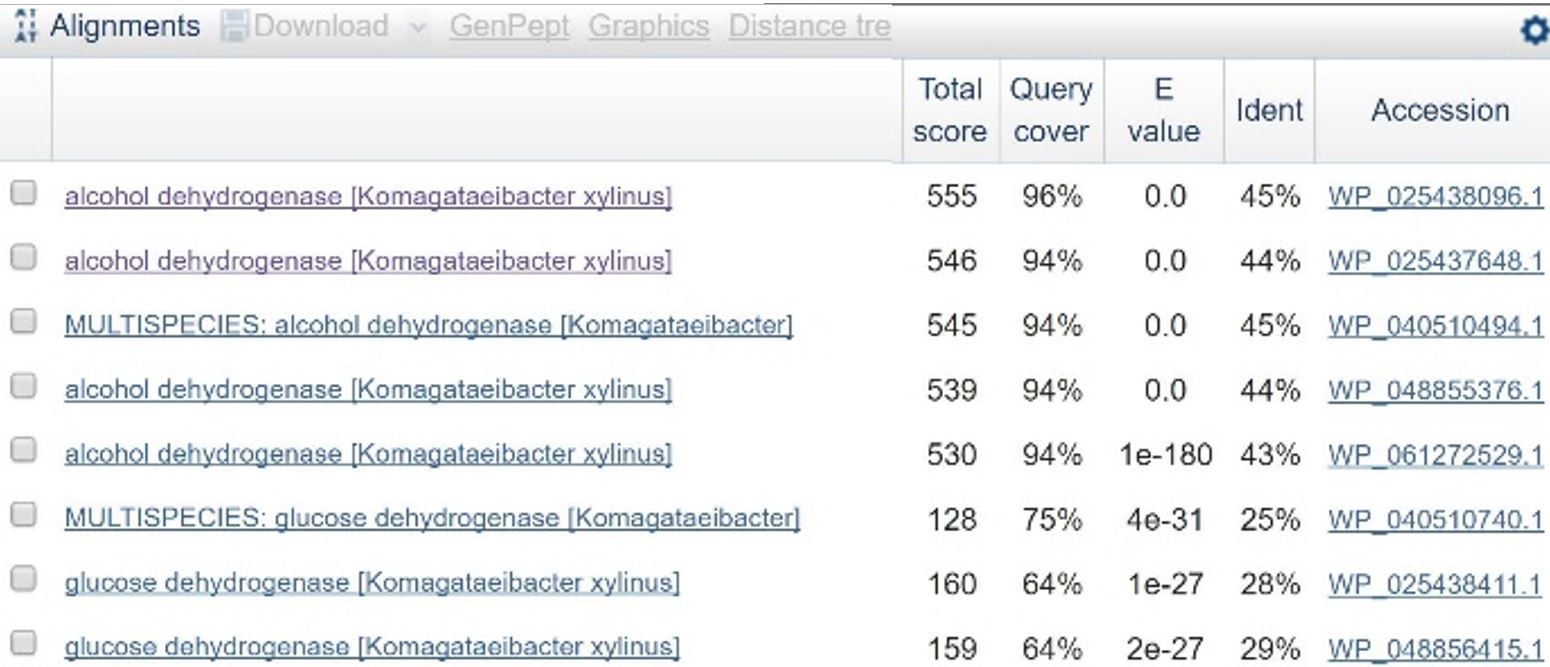
In order to design a construct increasing expression of PQQ-ADH and ALDH in <i>Ga. hansenii</i>, it was necessary to find the genome of the ATCC strain and identify the coding sequences for these genes. The whole genome shotgun sequence for our organism, ATCC 53582, is published on NCBI (Abbot, 2015) with annotations regarding the functions of specific sequences. Coding sequences are annotated with proposed gene products. Though there are several aldehyde dehydrogenase genes annotated in the genome, there is only one which is described as membrane-bound, matching the description from Mamlouk and Gullo (Mamlouk and Gullo, 2013). There are additionally multiple alcohol dehydrogenases. A known amino acid sequence for a homologous PQQ-ADH in <i>Comamonas testosteroni</i> was compared against sequences in the <i>Ga. hansenii</i> genome using BLAST (<b>Table 1</b>). One ADH enzyme found in the <i>Ga. hansenii</i> genome sequence matches the <i>C. testosteroni</i> sequence with a query cover value of 94% and an E value of 0 (third line of <b>Table 1</b>).</p> | In order to design a construct increasing expression of PQQ-ADH and ALDH in <i>Ga. hansenii</i>, it was necessary to find the genome of the ATCC strain and identify the coding sequences for these genes. The whole genome shotgun sequence for our organism, ATCC 53582, is published on NCBI (Abbot, 2015) with annotations regarding the functions of specific sequences. Coding sequences are annotated with proposed gene products. Though there are several aldehyde dehydrogenase genes annotated in the genome, there is only one which is described as membrane-bound, matching the description from Mamlouk and Gullo (Mamlouk and Gullo, 2013). There are additionally multiple alcohol dehydrogenases. A known amino acid sequence for a homologous PQQ-ADH in <i>Comamonas testosteroni</i> was compared against sequences in the <i>Ga. hansenii</i> genome using BLAST (<b>Table 1</b>). One ADH enzyme found in the <i>Ga. hansenii</i> genome sequence matches the <i>C. testosteroni</i> sequence with a query cover value of 94% and an E value of 0 (third line of <b>Table 1</b>).</p> | ||
<p><u>Creation of Golden Gate parts</u> | <p><u>Creation of Golden Gate parts</u> | ||
| − | + | ||
| − | + | ||
<p> | <p> | ||
In order to assemble the construct, the coding sequences for the genes of interest must be amplified from the <i>Ga. hansenii</i> genome and edited such that they have the correct Golden Gate overhangs and no internal BsaI or BsmBI restriction sites. The sequences were uploaded to Benchling for analysis and planning. The coding sequence for the membrane-bound ALDH contains a BsaI restriction site near the middle of the gene (<b>Figure 2</b>), and the PQQ-ADH coding sequence contains a BsmBI restriction site near the end of the gene (<b>Figure 3</b>). To eliminate the BsaI site in ALDH, primers were designed that would introduce a point mutation at the restriction site. One set of primers, igem2016_KOM_EtOH_01 and igem2016_KOM_EtOH_02, amplifies the sequence upstream of the restriction site, adding a type 3 Golden Gate prefix and removing the restriction site. Another set, igem2016_KOM_EtOH_03 and igem2016_KOM_EtOH_04, amplifies the region downstream of the restriction site, introducing a mutation to the site and adding a type 3 Golden Gate suffix to the end of the gene. These two products will be used in an overlap PCR reaction to create a final product with no BsaI restriction sites and the correct prefix and suffix for assembly. To remove the BsmBI site from the PQQ-ADH coding sequence, a set of primers (igem2016_KOM_EtOH_05 and igem2016_KOM_EtOH_06) was designed to amplify the region upstream of the restriction site and add a Golden Gate type 3 prefix to the beginning of the sequence. The reverse primer additionally adds a mutation to existing BsmBI restriction site and creates a new BsmBI restriction site that will be used to join the piece to a double-stranded DNA, igem2016_KOM_EtOH_07, containing the rest of the gene’s coding sequence appended with a Golden Gate type 3 suffix. The assembly of the PQQ-ADH part will therefore take place in two reactions: one reaction in which the upstream piece of DNA is created, and one reaction in which it is ligated to the gBlock. <b>Table 2</b> contains more information about each of these oligonucleotides. All were ordered from IDT.</p> | In order to assemble the construct, the coding sequences for the genes of interest must be amplified from the <i>Ga. hansenii</i> genome and edited such that they have the correct Golden Gate overhangs and no internal BsaI or BsmBI restriction sites. The sequences were uploaded to Benchling for analysis and planning. The coding sequence for the membrane-bound ALDH contains a BsaI restriction site near the middle of the gene (<b>Figure 2</b>), and the PQQ-ADH coding sequence contains a BsmBI restriction site near the end of the gene (<b>Figure 3</b>). To eliminate the BsaI site in ALDH, primers were designed that would introduce a point mutation at the restriction site. One set of primers, igem2016_KOM_EtOH_01 and igem2016_KOM_EtOH_02, amplifies the sequence upstream of the restriction site, adding a type 3 Golden Gate prefix and removing the restriction site. Another set, igem2016_KOM_EtOH_03 and igem2016_KOM_EtOH_04, amplifies the region downstream of the restriction site, introducing a mutation to the site and adding a type 3 Golden Gate suffix to the end of the gene. These two products will be used in an overlap PCR reaction to create a final product with no BsaI restriction sites and the correct prefix and suffix for assembly. To remove the BsmBI site from the PQQ-ADH coding sequence, a set of primers (igem2016_KOM_EtOH_05 and igem2016_KOM_EtOH_06) was designed to amplify the region upstream of the restriction site and add a Golden Gate type 3 prefix to the beginning of the sequence. The reverse primer additionally adds a mutation to existing BsmBI restriction site and creates a new BsmBI restriction site that will be used to join the piece to a double-stranded DNA, igem2016_KOM_EtOH_07, containing the rest of the gene’s coding sequence appended with a Golden Gate type 3 suffix. The assembly of the PQQ-ADH part will therefore take place in two reactions: one reaction in which the upstream piece of DNA is created, and one reaction in which it is ligated to the gBlock. <b>Table 2</b> contains more information about each of these oligonucleotides. All were ordered from IDT.</p> | ||
| + | </html> | ||
[[File:T--Austin UTexas--oligotable.png|thumb|left|'''Table 2''': Description of oligonucleotides ordered from IDT and their purposes. All of these are PCR primers except for igem2016_KOM_EtOH_07, which is a gBlock containing the end of the PQQ-ADH with a Golden Gate type 3 suffix appended.|500px]] | [[File:T--Austin UTexas--oligotable.png|thumb|left|'''Table 2''': Description of oligonucleotides ordered from IDT and their purposes. All of these are PCR primers except for igem2016_KOM_EtOH_07, which is a gBlock containing the end of the PQQ-ADH with a Golden Gate type 3 suffix appended.|500px]] | ||
| + | <html> | ||
<br><br> | <br><br> | ||
<center><a href="#top"><p>Back to Top</p></a></center> | <center><a href="#top"><p>Back to Top</p></a></center> | ||
Revision as of 21:19, 19 October 2016
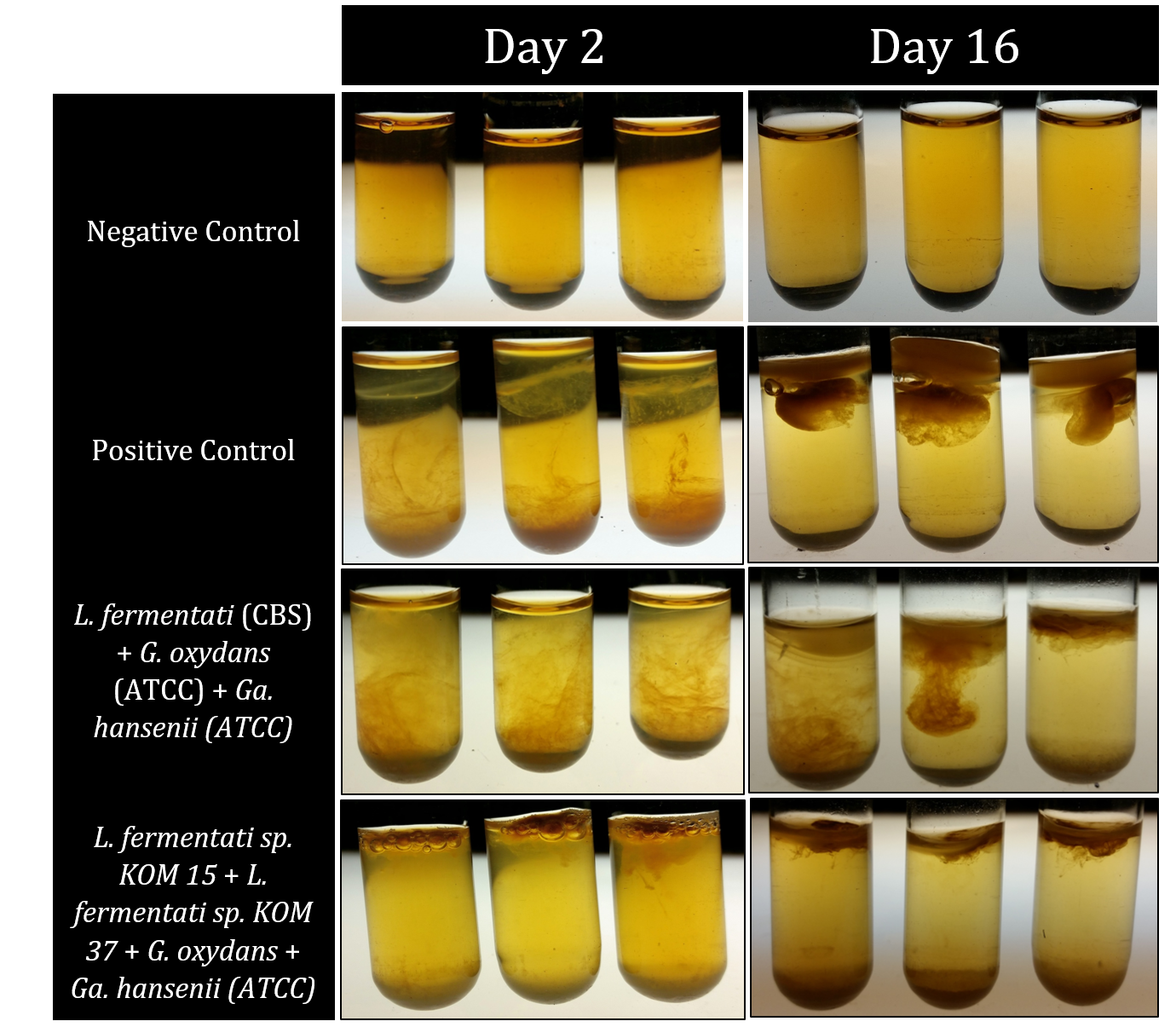
Figure 1: Shows samples from a series of completed recapitulation trials. A negative control triplicate set contained only tea media and experienced no microbial growth after 16 days. The positive control was inoculated with 600 uL of home-brewed kombucha and possessed distinct pellicle formation after only 2 days, forming a mature pellicle by Day 16. Both experimental trials pictured yielded successful recapitulations. Row 3 shows a set of trials that incorporated only microbes that had been purchased rather than isolated from kombucha itself. Row 4 shows successful recapitulations that contained two different strains of Lachancea fermentati each isolated from kombucha samples, as well as a strain of Gluconobacter oxydans and Gluconacetobacter hansenii. The cellulose pellicle produced in this set of trials is notably darker than the one observed for the purchased microbe strains as well as the positive controls.
Back to Top