Chelator Characteristics
As our project was to chelate dietary copper we searched for copper binding and storage proteins. We decided that the ideal copper chelator would have these properties:
- Should be able to bind multiple copper ions per peptide to increase the efficient use of cell resources.
- They should be from the prokaryotic domain because eukaryotic proteins can have expression issues in Escherichia coli.
- As E. coli naturally deals with copper toxicity by binding copper in the periplasm, periplasmic proteins may reduce toxicity to the host.
Using these criteria we found two copper chelators that we though would be useful. We designed gBlocks, codon optimised for E. coli, containing these parts both alone and linked to super-folder GFP. This form of the fluorescent protein was used because the standard GFP doesn’t fold particularly well in the periplasm where one of our chelators is intended to travel to. All the proteins, both alone and attached to sfGFP, were ordered with hexa-histidine tag so we could purify them. A concern was raised that the his tag would also weakly bind copper potentially affecting the results. However we decided that increasing the copper-binding would only enhance the protein.
Copper Storage Protein 1
Copper storage protein 1 is a protein discovered in an methane-oxidizing alphaproteobacterium called Methylosinus trichosporium OB3b. (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al* discovered Csp1 in 2015.
Csp1
Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins which are unstructured until they fold around metal ion clusters. They found an average copper affinity of approximately 1x10^-7M.
Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely as periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity.
We codon optimised Csp1 to E. coli and replaced the original TAT sequence with a TAT sequence from the E. coli protein CueO, which is also involved in copper regulation. To get Csp1 from the shipping vector to the pBAD expression system for testing the TAT sequence had to be modified but the addition of a serine residue after the initiator methionine. Serine was chosen over other amino acid possibilities because other TAT sequences seemed to have serine in this location. Unfortunately we were not able to demonstrate that Csp1 was transported to the periplasm in microscopy images.
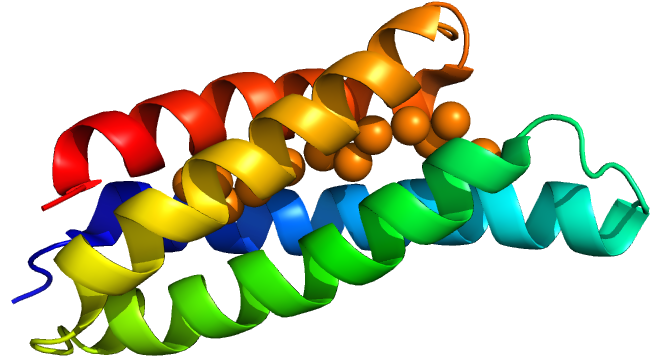
Csp1 sfGFP
Csp1 sfGFP is a form of Csp1 with a C terminal sfGFP tag. The sfGFP has a C terminal hexahistidine tag for purifications.
MymT
MymT
MymT is a small prokaryotic metallothein discovered in Mycobacterium tuberculosis. It can bind up to 7 Cu ions in a solvent-shielded core but apparently prefers 4-6. It is much smaller than Csp1 and exists as a monomer. It is cytoplasmic. No crystal structure exists but there is a structure of a eukaryotic homologue:

MymT sfGFP
MymT sfGFP is a form of MymT that has a C terminal sfGFP tag.

