(Blanked the page) |
|||
| (39 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <style> | ||
| + | |||
| + | /********************************* DEFAULT WIKI SETTINGS ********************************/ | ||
| + | |||
| + | #sideMenu, #top_title {display:none;} | ||
| + | #content { padding:0px; width:1000px; margin-top:-7px; margin-left:0px;} | ||
| + | body {background-color:white; } | ||
| + | #bodyContent h1, #bodyContent h2, #bodyContent h3, #bodyContent h4, #bodyContent h5 { margin-bottom: 0px; } | ||
| + | |||
| + | /********************************* MENU ********************************/ | ||
| + | /* Wrapper for the menu */ | ||
| + | .menu_wrapper { | ||
| + | width:150px; | ||
| + | height:100vh; | ||
| + | position:fixed; | ||
| + | padding:0px; | ||
| + | float:left; | ||
| + | background-color:#f2f2f2; | ||
| + | text-align:left; | ||
| + | } | ||
| + | |||
| + | /* styling for the menu items */ | ||
| + | .menu_item { | ||
| + | width:100%; | ||
| + | margin:-2px 0px 0px -20px; | ||
| + | padding: 10px 10px; | ||
| + | border-bottom: 1px solid #d3d3d3; | ||
| + | font-weight:bold; | ||
| + | color:#000000; | ||
| + | cursor: pointer; | ||
| + | } | ||
| + | |||
| + | /* when hovering on a menu item */ | ||
| + | .menu_item:hover { | ||
| + | color:#000000; | ||
| + | background-color: #72c9b6; | ||
| + | } | ||
| + | |||
| + | /* decoration icon for the menu buttons*/ | ||
| + | .icon { | ||
| + | float:right; | ||
| + | font-size:16px; | ||
| + | font-weight:bold; | ||
| + | } | ||
| + | |||
| + | /* this is the icon for when the content is collapsed */ | ||
| + | .plus::before { | ||
| + | content: "+"; | ||
| + | } | ||
| + | /* this is the icon for when the content is expanded */ | ||
| + | .less::before { | ||
| + | content: "–"; | ||
| + | } | ||
| + | |||
| + | /* styling for the ul that creates the buttons */ | ||
| + | ul.menu_items { | ||
| + | width:100%; | ||
| + | margin-left:0px; | ||
| + | padding:0px; | ||
| + | list-style: none; | ||
| + | } | ||
| + | |||
| + | /* styling for the li that are the menu items */ | ||
| + | .menu_items li { | ||
| + | width:90%; | ||
| + | margin-top:-2px; | ||
| + | padding: 15px 0px 15px 15px ; | ||
| + | display:block; | ||
| + | border-bottom: 1px solid #d3d3d3; | ||
| + | text-align:left; | ||
| + | font-weight:bold; | ||
| + | text-decoration:none; | ||
| + | color:#000000; | ||
| + | list-style-type:none; | ||
| + | cursor:pointer; | ||
| + | -webkit-transition: all 0.4s ease; | ||
| + | -moz-transition: all 0.4s ease; | ||
| + | -ms-transition: all 0.4s ease; | ||
| + | -o-transition: all 0.4s ease; transition: all 0.4s ease; | ||
| + | } | ||
| + | |||
| + | .menu_item a { | ||
| + | width: 100%; | ||
| + | margin-left: -20px; | ||
| + | padding: 11px 90px 12px 20px; | ||
| + | text-decoration: none; | ||
| + | color:black; | ||
| + | } | ||
| + | |||
| + | /* When hovering on a menu item */ | ||
| + | .menu_items li:hover { | ||
| + | background-color:#72c9b6; | ||
| + | color: #000000; | ||
| + | } | ||
| + | |||
| + | /* styling for the submenus */ | ||
| + | .submenu { | ||
| + | width:100%; | ||
| + | display: none; | ||
| + | font-weight:bold; | ||
| + | cursor:pointer; | ||
| + | |||
| + | } | ||
| + | |||
| + | /* moving the margin for the submenu ul list */ | ||
| + | ul.submenu { | ||
| + | width: 100%; | ||
| + | margin: 10px 0px -11px 0px; | ||
| + | list-style: none; | ||
| + | } | ||
| + | |||
| + | /*styling for the submenu buttons */ | ||
| + | .submenu li { | ||
| + | width: 100%; | ||
| + | margin-left: 10px; | ||
| + | margin-bottom: 0px; | ||
| + | } | ||
| + | |||
| + | |||
| + | /* hover state for the submenu buttons */ | ||
| + | .submenu li a { | ||
| + | width: 100%; | ||
| + | padding: 5px 10px; | ||
| + | display: inline-block; | ||
| + | border-bottom: 1px solid #d3d3d3; | ||
| + | background-color:white; | ||
| + | text-decoration:none; | ||
| + | color:#000000; | ||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | .submenu li a:hover { | ||
| + | background-color:#000000; | ||
| + | color: #72c9b6; | ||
| + | } | ||
| + | |||
| + | |||
| + | /* When the screen is smaller than 680px, the menu has the option to hide/show - this button controls that */ | ||
| + | .collapsable_menu_control { | ||
| + | width:100%; | ||
| + | padding: 15px 0px; | ||
| + | display:none; | ||
| + | background-color:#000000; | ||
| + | text-align:center; | ||
| + | font-weight:bold; | ||
| + | color:#72c9b6; | ||
| + | cursor:pointer; | ||
| + | -webkit-transition: all 0.4s ease; | ||
| + | -moz-transition: all 0.4s ease; | ||
| + | -ms-transition: all 0.4s ease; | ||
| + | -o-transition: all 0.4s ease; | ||
| + | transition: all 0.4s ease; | ||
| + | } | ||
| + | |||
| + | /* when hovering on that button */ | ||
| + | .collapsable_menu_control:hover { | ||
| + | background-color: #72c9b6; | ||
| + | color:#000000; | ||
| + | } | ||
| + | |||
| + | /********************************* CONTENT OF THE PAGE ********************************/ | ||
| + | |||
| + | /* Wrapper for the content */ | ||
| + | .content_wrapper { | ||
| + | width: 85%; | ||
| + | margin-left:150px; | ||
| + | padding:10px 0px; | ||
| + | float:left; | ||
| + | background-color:white; | ||
| + | } | ||
| + | |||
| + | /*LAYOUT */ | ||
| + | .column { | ||
| + | padding: 10px 0px; | ||
| + | float:left; | ||
| + | background-color:white; | ||
| + | } | ||
| + | |||
| + | .full_size { | ||
| + | width:100%; | ||
| + | } | ||
| + | |||
| + | .full_size img { | ||
| + | padding: 10px 15px; | ||
| + | width:96.5%; | ||
| + | } | ||
| + | |||
| + | .half_size { | ||
| + | width: 50%; | ||
| + | } | ||
| + | .half_size img { | ||
| + | padding: 10px 15px; | ||
| + | width: 93%; | ||
| + | } | ||
| + | |||
| + | |||
| + | .clear { | ||
| + | clear:both; | ||
| + | } | ||
| + | |||
| + | .highlight { | ||
| + | width: 90%; | ||
| + | margin: auto; | ||
| + | padding: 15px 5px; | ||
| + | background-color: #f2f2f2; | ||
| + | } | ||
| + | |||
| + | .judges-will-not-evaluate { | ||
| + | border: 4px solid #72c9b6; | ||
| + | display: block; | ||
| + | margin: 5px 15px; | ||
| + | width: 95%; | ||
| + | font-weight:bold; | ||
| + | } | ||
| + | |||
| + | |||
| + | /*STYLING */ | ||
| + | |||
| + | /* styling for the titles */ | ||
| + | .content_wrapper h1, .content_wrapper h2 { | ||
| + | padding:5px 15px; | ||
| + | border-bottom:0px; | ||
| + | color:#72c9b6; | ||
| + | |||
| + | } | ||
| + | .content_wrapper h3, .content_wrapper h4, .content_wrapper h5, .content_wrapper h6 { | ||
| + | padding:5px 15px; | ||
| + | border-bottom:0px; | ||
| + | color: #000000; | ||
| + | } | ||
| + | |||
| + | |||
| + | /* font and text */ | ||
| + | .content_wrapper p { | ||
| + | padding:0px 15px; | ||
| + | font-size: 13px; | ||
| + | font-family:Tahoma, Geneva, sans-serif; | ||
| + | } | ||
| + | |||
| + | /* Links */ | ||
| + | .content_wrapper a { | ||
| + | font-weight: bold; | ||
| + | text-decoration: underline; | ||
| + | text-decoration-color:#72c9b6; | ||
| + | color: #72c9b6; | ||
| + | -webkit-transition: all 0.4s ease; | ||
| + | -moz-transition: all 0.4s ease; | ||
| + | -ms-transition: all 0.4s ease; | ||
| + | -o-transition: all 0.4s ease; | ||
| + | transition: all 0.4s ease; | ||
| + | } | ||
| + | |||
| + | /* hover for the links */ | ||
| + | .content_wrapper a:hover { | ||
| + | text-decoration:none; | ||
| + | color:#000000; | ||
| + | } | ||
| + | |||
| + | /* non numbered lists */ | ||
| + | .content_wrapper ul { | ||
| + | padding:0px 20px; | ||
| + | font-size: 13px; | ||
| + | font-family:Tahoma, Geneva, sans-serif; | ||
| + | } | ||
| + | |||
| + | /* numbered lists */ | ||
| + | .content_wrapper ol { | ||
| + | padding:0px; | ||
| + | font-size: 13px; | ||
| + | font-family:Tahoma, Geneva, sans-serif; | ||
| + | } | ||
| + | |||
| + | /* Table */ | ||
| + | .content_wrapper table { | ||
| + | width: 97%; | ||
| + | margin:15px 10px; | ||
| + | border: 1px solid #d3d3d3; | ||
| + | border-collapse: collapse; | ||
| + | } | ||
| + | |||
| + | /* table cells */ | ||
| + | .content_wrapper td { | ||
| + | padding: 10px; | ||
| + | vertical-align: text-top; | ||
| + | border: 1px solid #d3d3d3; | ||
| + | border-collapse: collapse; | ||
| + | } | ||
| + | |||
| + | /* table headers */ | ||
| + | .content_wrapper th { | ||
| + | padding: 10px; | ||
| + | vertical-align: text-top; | ||
| + | border: 1px solid #d3d3d3; | ||
| + | border-collapse: collapse; | ||
| + | background-color:#f2f2f2; | ||
| + | } | ||
| + | |||
| + | /* Button class */ | ||
| + | .button_click { | ||
| + | margin: 10px auto; | ||
| + | padding: 15px; width:12%; | ||
| + | text-align:center; | ||
| + | font-weight:bold; | ||
| + | background-color: #72c9b6; | ||
| + | cursor:pointer; | ||
| + | -webkit-transition: all 0.4s ease; | ||
| + | -moz-transition: all 0.4s ease; | ||
| + | -ms-transition: all 0.4s ease; | ||
| + | -o-transition: all 0.4s ease; | ||
| + | transition: all 0.4s ease; | ||
| + | } | ||
| + | |||
| + | /* button class on hover */ | ||
| + | .button_click:hover { | ||
| + | background-color:#000000; | ||
| + | color:#72c9b6; | ||
| + | } | ||
| + | |||
| + | /********************************* RESPONSIVE STYLING ********************************/ | ||
| + | |||
| + | /* IF THE SCREEN IS LESS THAN 1000PX */ | ||
| + | |||
| + | @media only screen and (max-width: 1000px) { | ||
| + | |||
| + | #content {width:100%; } | ||
| + | .menu_wrapper {width:15%;} | ||
| + | .content_wrapper {margin-left: 15%;} | ||
| + | .menu_item {display:block;} | ||
| + | .icon {display:none;} | ||
| + | .highlight {padding:10px 0px;} | ||
| + | } | ||
| + | |||
| + | |||
| + | /* IF THE SCREEN IS LESS THAN 680PX */ | ||
| + | |||
| + | @media only screen and (max-width: 680px) { | ||
| + | .collapsable_menu_control { display:block;} | ||
| + | .menu_item {display:none;} | ||
| + | .menu_wrapper { width:100%; height: 15%; position:relative;} | ||
| + | .content_wrapper {width:100%; margin-left:0px;} | ||
| + | .column.half_size {width:100%; } | ||
| + | .column img { width: 100%; padding: 5px 0px;} | ||
| + | .icon {display:block;} | ||
| + | .highlight {padding:15px 5px;} | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--- THIS IS WHERE THE HTML BEGINS ---> | ||
| + | |||
| + | |||
| + | <!-- This tells the browser that your page is responsive --> | ||
| + | |||
| + | <head> | ||
| + | <meta name="viewport" content="width=device-width, initial-scale=1"> | ||
| + | </head> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="menu_wrapper" > | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="collapsable_menu_control"> MENU ▤ </div> | ||
| + | |||
| + | <ul id="accordion" class="accordion"> | ||
| + | |||
| + | <li class="menu_item"> <a href="https://2016.igem.org/Team:Nagahama">HOME </a> </li> | ||
| + | |||
| + | <li class="menu_item"> <div class="icon plus"></div> PROJECT | ||
| + | <ul class="submenu"> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Introduction"> ・Introduction </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Result and Discussion"> ・Result and Discussion </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Future"> ・Future </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Protocol"> ・Protocol </a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li class="menu_item"> <div class="icon plus"></div> BioBricks | ||
| + | <ul class="submenu"> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Our all BioBricks"> ・Our all BioBricks </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/BioBricks for medal"> ・BioBricks for medal </a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li class="menu_item"> <a href="https://2016.igem.org/Team:Nagahama/Safety"> Safety </a> </li> | ||
| + | |||
| + | |||
| + | <li class="menu_item"> <div class="icon plus"></div> Attributions | ||
| + | <ul class="submenu"> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Team"> ・Team</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Attributions"> ・Attributions </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Sponsors"> ・Sponsors</a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | <li class="menu_item"> <div class="icon plus"></div> Collaborations | ||
| + | <ul class="submenu"> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Collaborations"> ・Collaborations</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/The Annual Meeting Of The Genetics Society Of Japan"> ・The Annual Meeting Of The Genetics Society Of Japan </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Support Gifu"> ・Support Gifu</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/virginia"> ・Virginia</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Paris-Saclay"> ・Paris-Saclay</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Munich United"> ・Munich United</a></li> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li class="menu_item"> <div class="icon plus"></div> Human Practice | ||
| + | <ul class="submenu"> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Human Practice"> ・Human Practice </a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Contacting Local"> ・Contacting Local</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Contacting Japan"> ・Contacting Japan</a></li> | ||
| + | <li> <a href="https://2016.igem.org/Team:Nagahama/Intgrated Human Practice"> ・Intgrated Human Practice</a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="menu_item"> <a href="https://2016.igem.org/Team:Nagahama/Proof of concept"> Proof of concept </a> </li> | ||
| + | |||
| + | |||
| + | <li class="menu_item"> <a href="https://2016.igem.org/Team:Nagahama/Demonstrate your work"> Demonstrate your work </a> </li> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="content_wrapper"> | ||
| + | |||
| + | |||
| + | |||
| + | <h1 id="team_name"> </h1> | ||
| + | <h4 id="page_name"> </h4> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <script> | ||
| + | |||
| + | // This is the jquery part of your template. Try not modify any of this code since it makes your menu work. | ||
| + | |||
| + | $(document).ready(function() { | ||
| + | |||
| + | $("#HQ_page").attr('id',''); | ||
| + | |||
| + | |||
| + | if ( wgPageName.substring( 0, 8) == "Template") { // if the page is a template it displays the full name in a single line | ||
| + | $("#team_name").html( wgPageName ); | ||
| + | } | ||
| + | |||
| + | else if ( ( (wgPageName.match(/\//g) || []).length ) == 0 ) { // if it is the home page , just print the team's name | ||
| + | $("#team_name").html( wgPageName.substring( 5, wgPageName.length ) ); | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | // this adds the team's name as an h1 | ||
| + | $("#team_name").html( wgPageName.substring( 5 , wgPageName.indexOf("/") ) ); | ||
| + | |||
| + | // this adds the page's title as an h4 | ||
| + | $("#page_name").html ( ( wgPageName.substring( wgPageName.indexOf("/") + 1, wgPageName.length ) ).replace( /\/|_/g , " ") ); | ||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | $('#accordion').find('.menu_item').click(function(){ | ||
| + | |||
| + | //Expand or collapse this panel | ||
| + | submenu = $(this).find('.submenu'); | ||
| + | submenu.toggle(); | ||
| + | |||
| + | icon = $(this).find('.icon'); | ||
| + | |||
| + | if ( !$( submenu ).is(':visible') ) { | ||
| + | icon.removeClass("less").addClass("plus"); | ||
| + | } | ||
| + | else { | ||
| + | icon.removeClass("plus").addClass("less"); | ||
| + | } | ||
| + | |||
| + | //Hide the other panels | ||
| + | $(".submenu").not(submenu).hide(); | ||
| + | $(".icon").not(icon).removeClass("less").addClass("plus"); | ||
| + | }); | ||
| + | |||
| + | |||
| + | $(".collapsable_menu_control").click(function() { | ||
| + | $(".menu_item").toggle(); | ||
| + | }); | ||
| + | |||
| + | $( window ).resize(function() { | ||
| + | $(".menu_item").show(); | ||
| + | }); | ||
| + | |||
| + | |||
| + | }); | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </script> | ||
| + | |||
| + | |||
| + | </html> | ||
| + | BioBrick Parts to achieve each medal requirement | ||
| + | |||
| + | Bronze | ||
| + | <br/> | ||
| + | We created 3 new BioBricks [1 ](https://2016.igem.org/Team:Nagahama/Our_all_BioBricks) for bronze medal criterion. And have documented and submitted them to igem Registry. | ||
| + | We introduce yajo (http://parts.igem.org/Part:BBa_K1950005) of them. This biobrick is Key BioBrics in “Flavorator” project. Yajo encodes novel 1-deoxyxylulose-5-phosphate synthase. The novel 1-deoxyxylulose-5-phosphate synthase is able to synthesize 1-deoxy-D-xylulose 5-phosphate (DXP) from ribulose 5-phosphate, proving a novel route from a pentose phosphate to DXP Therefor the enzyme can catalyze in the polyisoprenoid biosynthetic pathway. | ||
| + | |||
| + | Silver | ||
| + | <br/> | ||
| + | We created 3 new BioBrick devices for silver medal criterion. | ||
| + | <br/> | ||
| + | 1.trc promoter + ispC-ribB(G108S) fusion | ||
| + | <br/> | ||
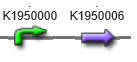
| + | We want to make Escherichia coli synthesize a lot of farnesol, which is one kind of terpenes. We can achieve this purpose by fusing two genes, ispC and ribB(G108S).(http://parts.igem.org/Part:BBa_K1950010) | ||
| + | <br/><br/> | ||
| + | [[File:パーツ10.png|800px|thumb|center|fig.1:Part:BBa_K1950010]] | ||
| + | <br/> | ||
| + | We tested whether the fusing gene was made by measuring ispC and ribB(G108S) transcription. The amounts of transcription were quantified by RT-RCR. In the fusion gene, ispC and ribB(G108S) are transferred from a single promoter. If two genes are fused successfully, two genes are expected to be higher in the recombinant than in WT. | ||
| + | <br/> | ||
| + | 2-1:PgpB.dev | ||
| + | <br/> | ||
| + | 2-2:YbjG.dev | ||
| + | <br/> | ||
| + | To realize "Flovorator" we need to make E. coli produce Farnesol. | ||
| + | We have succeeded in confirming amount of transcription of PgpB.dev and YbjG.dev . | ||
| + | Our new biobrick device “PgpB.dev” and ”YbjG.dev” worked as expected. | ||
| + | Because we validate this fact by RT-PCR analysis experimentally. We have documented and submitted this new Biobrick device (http://parts.igem.org/Part:BBa_K1950007) | ||
| + | (http://parts.igem.org/Part:BBa_K1950008) to iGEM Registry . | ||
| + | <br/> | ||
| + | [[File:遺伝学会パーツS1.png|800px|thumb|fig.2:Part:BBa_K1950007_fig.3:Part:BBa_K1950008]] | ||
| + | <br/> | ||
| + | We want to make the E. coli produces farnesol which are one of the terpenes. | ||
| + | To produce great quantity of terpenes they need many terpene precursor and Farnesol synthase. | ||
| + | E. coli produces a small amount of the terpene precursor in MEP pathway. | ||
| + | In addition, E. coli has dephosphorylation enzyme of endogenous. | ||
| + | This is likely to synthesize farnesol from Farnesyl diphosphate (FPP), which is a precursor of farnesol. | ||
| + | In MEP pathway, there are four enzymes (ispD, ispF, idi, dxs) which are speed limiting enzyme for terpenes precursors produce in E. coli. | ||
| + | Last year, in order to, create a high-yield strains producing IPP and DMAPP, we exogenously engineered to superimpose these genes into E. coli to create strains overproducing IPP and DMAPP. | ||
| + | This year, in order to generate a large amount of further farnesol, exogenously engineer genes coding for farnesol enzyme synthesis , YbjG and PgpB, as farnesol synthesis in E. coli. These are endogenous dephosphorylation enzyme of E. coli. | ||
| + | Farnesol is generated through FPP hydrolysis by PgpB and YbjG of endogenous phosphatases , which are induced by an increased intracellular FPP level. | ||
| + | To confirm whether the transcription of these genes is going well or not, we confirmed amount of the transcription of these genes by RT-PCR. | ||
| + | |||
| + | Result:(fig◯) | ||
| + | |||
| + | Discussion: | ||
| + | <br> | ||
| + | [[File:Photo.jpg|800px|thumb|center|Fig4.The result of RT-PCR ybjG pgpB Fusion negative control]] | ||
| + | <br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | |||
| + | Gold | ||
| + | <br/> | ||
| + | marA :The activator of AcrAB-TolC multidrug efflux pump exports some terpenes | ||
| + | We improved the characterization of a previously existing BioBrick Part BBa_K1230000 and (http://parts.igem.org/Part:BBa_K1230000) the characterization of a previously existing BioBrick marAgene as BBa_K1653006 . In existing part's information of marA, it gives E. coli resistance against kanamycin and give E-Coli resistance against geraniol as one of the terpene and decrease its intracellular concentration. In this year, we confirmed that overexpressing of marA gives E. coli resistance against farnesol as one of the terpene. This information is very beneficial for other iGEMers to production of toxic organic substances that are produced using bacteria. | ||
| + | We introduce an activator gene of AcrAB-TolC efflux pump (MarA) to release the farnesol from the cells and increase the content in the media that shows increase these flavors in the "Flavolator". In our study, we confirmed that overexpressing of marA gives host E. coli high resistance against farnesol. | ||
| + | <br/> | ||
| + | [[File:E.colimarA result .jpg|800px|thumb|center|Fig.5:Colony formation of E. coli JM109 engineered with marA on farnesol overlaid plates. | ||
| + | <br> | ||
| + | |||
| + | ''E.coli'' JM109 and ''E.coli'' JM109 (marA) were dropped on LBGMg agar plates in serial ten-fold dilutions (10⁻¹~〖10〗^(-6)), overlaid with 30 % (v/v) farnesol hexane solution (farnesol solution) and incubated at 30°C for 24 h. This figure shows that ''E.coli'' JM109 (marA) cells that overexpress the marA product better than the control ''E.coli'' JM109 wild type cells survived plates overloyed by 30 % farnesol solution. | ||
| + | ]] | ||
| + | <br/> | ||
| + | Farnesol production device(http://parts.igem.org/Part:BBa_K1653025) | ||
| + | |||
| + | To realize "Flovorator" we need to make E. coli produce Farnesol. Surprisingly, last year, we have succeeded in farnesol production by Farnesol production device. Our previous biobrick device "Farnesol production device" worked as expected. Because we validated this fact by GC-MS analysis experimentally. | ||
| + | We confirmed expression level of genes used in the 2015 fiscal year project for reference of farnesol production increase. | ||
| + | We improved the characterization of a previously existing BioBrick Part BBa_K1653025 | ||
| + | In existing part's information of farnesol production device, it gives E. coli farnesol production. In this year, | ||
| + | |||
| + | This information is very beneficial for other iGEMers to farnesol that are produced using bacteria. | ||
| + | |||
| + | [[File:遺伝学会パーツS2.png|800px|thumb|fig.6:Part:BBa_K1653025]] | ||
| + | <br/> | ||
| + | <br/><br/><br/><br/><br/><br/><br/> | ||
| + | 結果の図を入れる | ||
Latest revision as of 01:50, 20 October 2016
BioBrick Parts to achieve each medal requirement
Bronze
We created 3 new BioBricks [1 ](https://2016.igem.org/Team:Nagahama/Our_all_BioBricks) for bronze medal criterion. And have documented and submitted them to igem Registry.
We introduce yajo (http://parts.igem.org/Part:BBa_K1950005) of them. This biobrick is Key BioBrics in “Flavorator” project. Yajo encodes novel 1-deoxyxylulose-5-phosphate synthase. The novel 1-deoxyxylulose-5-phosphate synthase is able to synthesize 1-deoxy-D-xylulose 5-phosphate (DXP) from ribulose 5-phosphate, proving a novel route from a pentose phosphate to DXP Therefor the enzyme can catalyze in the polyisoprenoid biosynthetic pathway.
Silver
We created 3 new BioBrick devices for silver medal criterion.
1.trc promoter + ispC-ribB(G108S) fusion
We want to make Escherichia coli synthesize a lot of farnesol, which is one kind of terpenes. We can achieve this purpose by fusing two genes, ispC and ribB(G108S).(http://parts.igem.org/Part:BBa_K1950010)
We tested whether the fusing gene was made by measuring ispC and ribB(G108S) transcription. The amounts of transcription were quantified by RT-RCR. In the fusion gene, ispC and ribB(G108S) are transferred from a single promoter. If two genes are fused successfully, two genes are expected to be higher in the recombinant than in WT.
2-1:PgpB.dev
2-2:YbjG.dev
To realize "Flovorator" we need to make E. coli produce Farnesol.
We have succeeded in confirming amount of transcription of PgpB.dev and YbjG.dev .
Our new biobrick device “PgpB.dev” and ”YbjG.dev” worked as expected.
Because we validate this fact by RT-PCR analysis experimentally. We have documented and submitted this new Biobrick device (http://parts.igem.org/Part:BBa_K1950007)
(http://parts.igem.org/Part:BBa_K1950008) to iGEM Registry .
We want to make the E. coli produces farnesol which are one of the terpenes.
To produce great quantity of terpenes they need many terpene precursor and Farnesol synthase.
E. coli produces a small amount of the terpene precursor in MEP pathway.
In addition, E. coli has dephosphorylation enzyme of endogenous.
This is likely to synthesize farnesol from Farnesyl diphosphate (FPP), which is a precursor of farnesol.
In MEP pathway, there are four enzymes (ispD, ispF, idi, dxs) which are speed limiting enzyme for terpenes precursors produce in E. coli.
Last year, in order to, create a high-yield strains producing IPP and DMAPP, we exogenously engineered to superimpose these genes into E. coli to create strains overproducing IPP and DMAPP.
This year, in order to generate a large amount of further farnesol, exogenously engineer genes coding for farnesol enzyme synthesis , YbjG and PgpB, as farnesol synthesis in E. coli. These are endogenous dephosphorylation enzyme of E. coli.
Farnesol is generated through FPP hydrolysis by PgpB and YbjG of endogenous phosphatases , which are induced by an increased intracellular FPP level.
To confirm whether the transcription of these genes is going well or not, we confirmed amount of the transcription of these genes by RT-PCR.
Result:(fig◯)
Discussion:
Gold
marA :The activator of AcrAB-TolC multidrug efflux pump exports some terpenes
We improved the characterization of a previously existing BioBrick Part BBa_K1230000 and (http://parts.igem.org/Part:BBa_K1230000) the characterization of a previously existing BioBrick marAgene as BBa_K1653006 . In existing part's information of marA, it gives E. coli resistance against kanamycin and give E-Coli resistance against geraniol as one of the terpene and decrease its intracellular concentration. In this year, we confirmed that overexpressing of marA gives E. coli resistance against farnesol as one of the terpene. This information is very beneficial for other iGEMers to production of toxic organic substances that are produced using bacteria.
We introduce an activator gene of AcrAB-TolC efflux pump (MarA) to release the farnesol from the cells and increase the content in the media that shows increase these flavors in the "Flavolator". In our study, we confirmed that overexpressing of marA gives host E. coli high resistance against farnesol.
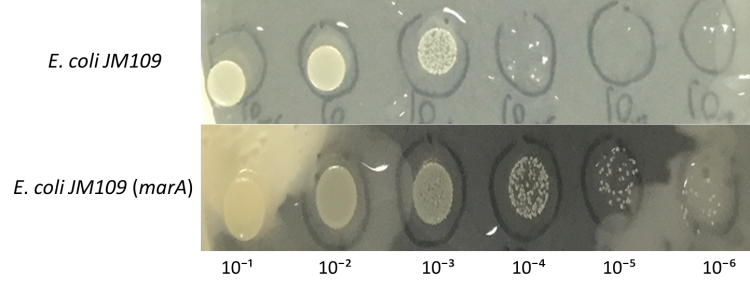
E.coli JM109 and E.coli JM109 (marA) were dropped on LBGMg agar plates in serial ten-fold dilutions (10⁻¹~〖10〗^(-6)), overlaid with 30 % (v/v) farnesol hexane solution (farnesol solution) and incubated at 30°C for 24 h. This figure shows that E.coli JM109 (marA) cells that overexpress the marA product better than the control E.coli JM109 wild type cells survived plates overloyed by 30 % farnesol solution.
Farnesol production device(http://parts.igem.org/Part:BBa_K1653025)
To realize "Flovorator" we need to make E. coli produce Farnesol. Surprisingly, last year, we have succeeded in farnesol production by Farnesol production device. Our previous biobrick device "Farnesol production device" worked as expected. Because we validated this fact by GC-MS analysis experimentally. We confirmed expression level of genes used in the 2015 fiscal year project for reference of farnesol production increase. We improved the characterization of a previously existing BioBrick Part BBa_K1653025 In existing part's information of farnesol production device, it gives E. coli farnesol production. In this year,
This information is very beneficial for other iGEMers to farnesol that are produced using bacteria.
結果の図を入れる