| (21 intermediate revisions by 3 users not shown) | |||
| Line 141: | Line 141: | ||
padding-bottom: 5px; | padding-bottom: 5px; | ||
} | } | ||
| − | |||
| − | |||
| − | |||
| Line 151: | Line 148: | ||
</head> | </head> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 195: | Line 186: | ||
<a href="https://2016.igem.org/Team:BostonU_HW/Demonstrate">Demonstration</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Demonstrate">Demonstration</a> | ||
<a href="https://2016.igem.org/Team:BostonU_HW/Proof">Proof</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Proof">Proof</a> | ||
| − | <a href="https://2016.igem.org/Team:BostonU_HW/Design"> | + | <a href="https://2016.igem.org/Team:BostonU_HW/Design">Design</a> |
<a href="https://2016.igem.org/Team:BostonU_HW/Parts">Parts</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Parts">Parts</a> | ||
</div> | </div> | ||
| Line 213: | Line 204: | ||
<a href="https://2016.igem.org/Team:BostonU_HW/HP/Silver">Silver</a> | <a href="https://2016.igem.org/Team:BostonU_HW/HP/Silver">Silver</a> | ||
<a href="https://2016.igem.org/Team:BostonU_HW/HP/Gold">Gold</a> | <a href="https://2016.igem.org/Team:BostonU_HW/HP/Gold">Gold</a> | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</li> | </li> | ||
| Line 220: | Line 210: | ||
<a href="#" class="dropbtn">AWARDS</a> | <a href="#" class="dropbtn">AWARDS</a> | ||
<div class="dropdown-content"> | <div class="dropdown-content"> | ||
| − | <a href="https://2016.igem.org/Team:BostonU_HW/ | + | <a href="https://2016.igem.org/Team:BostonU_HW/Integrated_Practices">Integrated Practices</a> |
<a href="https://2016.igem.org/Team:BostonU_HW/Software">Software</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Software">Software</a> | ||
</div> | </div> | ||
| Line 230: | Line 220: | ||
<a href="https://2016.igem.org/Team:BostonU_HW/Silver">Silver</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Silver">Silver</a> | ||
<a href="https://2016.igem.org/Team:BostonU_HW/Gold">Gold</a> | <a href="https://2016.igem.org/Team:BostonU_HW/Gold">Gold</a> | ||
| − | + | ||
</div> | </div> | ||
</li> | </li> | ||
| Line 377: | Line 367: | ||
<div class="col-md-10" style="font-size: 1.5em; line-height: 130%"> | <div class="col-md-10" style="font-size: 1.5em; line-height: 130%"> | ||
| − | In our Silver Medal Human Practices page, we outlined our public outreach and industry visits. These experiences drew our team to focus on accessibility in synthetic biology as a theme moving forward with our human practice contributions. For Silver HP, we contributed a set of informational blog posts on the history of intellectual property, and IP in software and in synthetic biology today. We welcome you to read these at our WordPress site> | + | In our Silver Medal Human Practices page, we outlined our public outreach and industry visits. These experiences drew our team to focus on accessibility in synthetic biology as a theme moving forward with our human practice contributions. For Silver HP, we contributed a set of informational blog posts on the history of intellectual property, and IP in software and in synthetic biology today. We welcome you to read these at our WordPress site. |
| + | <br><br> | ||
| + | <a href="https://buigem2016.wordpress.com/" style="padding-right:20px"> <button type="button" class="btn btn-primary" id="down">WORDPRESS BLOG</button></a> | ||
<br><br> | <br><br> | ||
Moving forward with the development of Neptune, we decided that we would extend our silver medal HP theme of accessibility in synthetic biology. Indeed, we were developing a complete, end-to-end microfluidic development workflow. Having seen how inaccessible and prohibitively costly microfluidics are for researchers, and also having studied the virtues of open source tools for synthetic biology, we decided to integrate accessibility into our implementation of Neptune. | Moving forward with the development of Neptune, we decided that we would extend our silver medal HP theme of accessibility in synthetic biology. Indeed, we were developing a complete, end-to-end microfluidic development workflow. Having seen how inaccessible and prohibitively costly microfluidics are for researchers, and also having studied the virtues of open source tools for synthetic biology, we decided to integrate accessibility into our implementation of Neptune. | ||
| Line 383: | Line 375: | ||
In this page, we cover 3 ways in which we expand on and integrate the theme of accessibility to our final product, Neptune. | In this page, we cover 3 ways in which we expand on and integrate the theme of accessibility to our final product, Neptune. | ||
<br> | <br> | ||
| − | + | <ol> | |
| − | < | + | <li>we made it a project criteria that Neptune must interface with low cost, open and readily available tools and hardware to create microfluidics</li> |
| − | + | <li>we partnered with the NONA Research Foundation, an organization dedicated to increasing access, collaboration, and building a community around synthetic biology software tools</li> | |
| − | < | + | <li>we offer our team as a point of contact to other iGEM teams that have created software solutions and would like to have these tools protected and stored on NONA</li> |
| − | + | </ol> | |
</div> | </div> | ||
| Line 405: | Line 397: | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: | + | <div style="font-size: 3em; line-height: 150%;"> Neptune: Low cost, easy to use, accessible </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 411: | Line 403: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | + | ||
| + | As we developed Neptune, we were faced with design choices that would have a large impact on how our workflow integrates with the synthetic biology community. In light of our Silver HP discoveries, we decided the most important design choice would be to make our workflow accessible to all: we want a workflow so easy to use that researchers in synthetic biology will be excited to make microfluidic chips. We want a workflow that interfaces with fabrication protocols that are low cost, such that even DIY hobbyists could make new chips. We wanted a workflow whose hardware could be fabricated with a 3D printer, and whose software is open source so that the entire synthetic biology community can contribute to new features in Neptune. Further, we wanted to ensure our workflow completely and cleanly integrates with current fabrication methods, so that we are still following current standards. | ||
| + | |||
| + | |||
<br><br> | <br><br> | ||
</div> | </div> | ||
| Line 424: | Line 419: | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> Open Source Software </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 431: | Line 426: | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | |||
| − | |||
| − | + | While this is already an iGEM requirement, we are proud to say our source code for Neptune is licensed as open under a BSD-II license. Further, we are very proud to say that Neptune is public on GitHub, and we encourage developers to fork the repo. | |
| − | + | <br><br> | |
| − | + | Neptune is free to download from GitHub too, and very soon NONA will provide a distribution method. | |
| − | + | <br><br> | |
| − | + | <a href="https://github.com/CIDARLAB/Neptune" style="padding-right:20px"> <button type="button" class="btn btn-primary" id="down">GITHUB REPOSITORY</button></a> | |
| + | <a href="http://nonasoftware.org/available-software/" style="padding-right:20px"> <button type="button" class="btn btn-primary" id="down">NONA</button></a> | ||
| + | |||
| + | |||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| + | |||
| + | <br><br><br> | ||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-1"> | ||
| + | </div> | ||
| + | <div class="col-md-11"> | ||
| + | <div style="font-size: 2em; line-height: 150%;"> Simple Design Process </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="row" style="padding-top:20px; padding-bottom:20px;"> | ||
| + | <div class="col-md-1"></div> | ||
| + | <div class="col-md-10"> | ||
| + | |||
| + | Accessibility means ease of use. Ease of use means simplicity; it means a no-brainer user interface and powerful code to help you design microfluidics with ease. Indeed, the biggest setback to designing microfluidics before Neptune was the tedious, long, and difficult process of hand-specifying all features in the microfluidic device. In Neptune, simple english-language descriptions allow you to specify complex microfluidic designs. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="col-md-1"></div> | ||
| + | </div> | ||
<div class="row"> | <div class="row"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
<div class="row"> | <div class="row"> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="https://static.igem.org/mediawiki/2016/d/ | + | <img src="https://static.igem.org/mediawiki/2016/d/d0/T--BostonU_HW--lfr_priyak.jpg" width="125%"> |
</div> | </div> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/a/a0/T--BostonU_HW--chipk2_priyak.jpg" width="75%"> |
</div> | </div> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/f/f7/T--BostonU_HW--chipk1_priyak.jpg" width="75%"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 460: | Line 477: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| − | |||
<br><br><br> | <br><br><br> | ||
| − | + | ||
| + | <div class="row"> | ||
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> Tutorials, Wiki's, Documentation </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 474: | Line 491: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | + | ||
| − | + | Not knowing how to use a new tool can dissuade researchers from ever bothering to learn; poor documentation, lack of support for new users, confusing or nonexistent tutorials- all of these add up to a big headache to a researcher who wants to get into microfluidics. | |
| − | + | <br><br> | |
| − | + | We targeted this issue by creating a tutorial video for our software and hardware workflow. This tutorial is a complete “Hello World” walkthrough that will guide new researchers step-by-step through the microfluidic creation process. We hope that this will encourage anyone who had been intimidated by microfluidic design to come forward and give it a shot. | |
| − | We | + | <br><br> |
| − | + | A tutorial video is good, but not complete. We followed up by starting a extensive written tutorial to writing LFR and MINT files (to design microfluidics,) and written tutorials on how to setup the hardware components. We also added wiki into to the GitHub repositories for Neptune and our hardware, providing yet another resource for new users to try. | |
| + | |||
| + | |||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| − | |||
| − | |||
<div class="row"> | <div class="row"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| Line 490: | Line 507: | ||
<div class="row"> | <div class="row"> | ||
<div calss="col-md-4"> | <div calss="col-md-4"> | ||
| − | <img src=" | + | <img src="" width="75%"> |
</div> | </div> | ||
<div calss="col-md-4"> | <div calss="col-md-4"> | ||
| − | <img src=" | + | <img src="" width="75%"> |
</div> | </div> | ||
<div calss="col-md-4"> | <div calss="col-md-4"> | ||
| − | <img src=" | + | <img src="" width="75%"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 503: | Line 520: | ||
</div> | </div> | ||
| − | + | <br><br><br> | |
| − | + | <div class="row"> | |
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> Integrated, Low Cost Fabrication Protocols </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 516: | Line 533: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | + | ||
| + | This is truly where the Gold Medal HP contributions become apparent. We had a clear choice here: We could have made Neptune interface with industry grade, powerful and very expensive fabrication methods using photolithography and plasma bonding. If we decided to do this, there would be a blunt eighty-thousand-dollar barrier of entry to researchers and hobbyists looking to use Neptune. That would be unacceptable. We could have had Neptune’s microfluidics be controlled using very high precision, high accuracy pumps that would provide flawless flow rates. But the hardware and infrastructure for these pumps would cost hundreds of dollars per pump, and the cost would not scale. We could not settle for this. | ||
<br><br> | <br><br> | ||
| − | + | Instead, we framed our workflow around the low cost MakerFluidics fabrication protocol. In this protocol, the eighty thousand dollar fabrication process of photolithography is bypassed; instead our microfluidic chips are fabricated with a simple CNC mill, costing a order of magnitude less, while still delivering high performance devices. | |
| − | + | <br><br> | |
| − | + | We made it our project goal, to fully integrate accessibility into Neptune. Neptune is microfluidics for industry, for research, for education and for hobbyists. We made Neptune have the lowest possible bar to entry; it is the only microfluidic design, fabrication and control workflow all integrated into one software tool-- and the best part is it can be used by anyone. We made accessibility the centerpiece of Neptune. | |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | + | <div class="row"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <div class="row"> | + | |
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
<div class="row"> | <div class="row"> | ||
| − | + | <div class="col-md-3"> | |
| − | + | </div> | |
| − | + | <div class="col-md-4"> | |
| − | <div | + | <img src="https://static.igem.org/mediawiki/2016/2/28/T--BostonU_HW--make.png" width="175%"> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <img src="https://static.igem.org/mediawiki/2016/2/ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | <div class="col-md-4"> | ||
| + | </div> | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
| Line 570: | Line 563: | ||
<br><br><br> | <br><br><br> | ||
| − | + | <div class="row"> | |
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> Open Hardware Schematics </div> |
</div> | </div> | ||
</div> | </div> | ||
<div class="row" style="padding-top:20px; padding-bottom:20px;"> | <div class="row" style="padding-top:20px; padding-bottom:20px;"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| − | <div class="col-md-10"> | + | <div class="col-md-10"> |
| − | + | A big aspect of enabling accessible microfluidics is reducing the barrier of entry for controlling these devices. Microfluidics are controlled by pumps that generate pressures gradients to push or pull fluids, or open and close valves. Most modern microfluidic control pumps are expensive and difficult to integrate, making it hard to scale for larger microfluidic designs, and dissuasive for hobbyists. | |
<br><br> | <br><br> | ||
| − | + | We chose to directly target our hardware designs to this problem. We developed completely open 3D print design schematics for our microfluidic control infrastructure. These designs are available for download on our GitHub repo, and server though Neptune as well! Furthermore, we developed a completely original hardware design to control syringe pumps using very low cost servo motors. | |
| − | < | + | |
| − | + | </div> | |
| − | + | <div class="col-md-1"></div> | |
</div> | </div> | ||
| − | + | <div class="row"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <div class="row"> | + | |
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
<div class="row"> | <div class="row"> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="" width=" | + | <img src="https://static.igem.org/mediawiki/2016/9/9b/T--BostonU_HW--hardwarek_priyak.jpg" width="75%"> |
</div> | </div> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="" width=" | + | <img src="https://static.igem.org/mediawiki/2016/3/3b/T--BostonU_HW--hardwarek2_priyak.jpg" width="75%"> |
</div> | </div> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src=" | + | <img src="https://static.igem.org/mediawiki/2016/b/b6/T--BostonU_HW--hardwarek3_priyak.jpg" width="75%"> |
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 613: | Line 599: | ||
</div> | </div> | ||
| − | |||
| − | <div class="row"> | + | <br><br><br><br><br><br> |
| + | |||
| + | |||
| + | <div class="row"> | ||
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: | + | <div style="font-size: 3em; line-height: 150%;">NONA Research Foundation Partnership </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 625: | Line 613: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | + | ||
| + | Integrating our workflow with the idea of accessibility was a huge step; we successfully build Neptune as the most accessible microfluidics creation tool to date, with low cost fabrication protocols, open hardware designs, and powerful design algorithms- virtually any research lab can now realistically consider microfluidics as an option, without devoting huge costs or needing expertise in the field. | ||
<br><br> | <br><br> | ||
| − | + | A big step, but we had to go further. The biggest issue we faced as the summer drew to a close was sustainability. Indeed, a software tool is only accessible if it is properly maintained, updated and if it has a community supporting it. What would happen to Neptune after iGEM? Where will Neptune be in 5 years? If we seriously cared about promoting an accessible workflow, we had to make sure we considered the future of our tool. | |
| − | + | ||
| + | |||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| Line 635: | Line 625: | ||
| − | + | <div class="row"> | |
| − | + | ||
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> What is NONA? </div> |
</div> | </div> | ||
</div> | </div> | ||
<div class="row" style="padding-top:20px; padding-bottom:20px;"> | <div class="row" style="padding-top:20px; padding-bottom:20px;"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| − | <div class="col-md-10"> | + | <div class="col-md-10"> |
| − | + | And so we proceeded to reach out to the NONA Research Foundation, a nonprofit whose aim is to broaden access to and develop a community around open source synthetic biology software tools. As cited from the organization, “Nona creates long-lasting solutions to the challenges posed by the rapidly growing complexity of software in the synthetic biology community by using the power of open source.” | |
| − | + | <br><br> | |
| − | + | Integrating with NONA provided 3 long-term, sustainable solutions to promoting the accessibility of Neptune and microfluidics to the synthetic biology community. | |
| + | <br><br> | ||
| + | 1) NONA would distribute Neptune, allowing Neptune to have a “home” where is can easily be found (beyond GitHub.) | ||
| + | <br><br> | ||
| + | 2) NONA is dedicated to integrating all open synthetic biology software tools under one umbrella, thus building a community of researchers and developers who who will use and support these tools. Thus, NONA will be a medium through which other developers can contribute to Neptune, and NONA will be a place researchers can go to learn more about how to use Neptune, ask for help, and so forth. | ||
| + | <br><br> | ||
| + | 3) Overall, by having a dedicated community and website where Neptune will always have a place, we are making it very hard for Neptune to be abandoned. Quite the opposite, with NONA will ensure Neptune is sustained for years to come, while also providing the opportunity for a community to flourish around Neptune and similar software tools. | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <br><br><br> | ||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-1"> | ||
| + | </div> | ||
| + | <div class="col-md-11"> | ||
| + | <div style="font-size: 2em; line-height: 150%;"> Passing Neptune to NONA </div> | ||
| + | </div> | ||
</div> | </div> | ||
| + | <div class="row" style="padding-top:20px; padding-bottom:20px;"> | ||
| + | <div class="col-md-1"></div> | ||
| + | <div class="col-md-10"> | ||
| − | <div class="row"> | + | We met with members of NONA to discuss integrating Neptune with their foundation. Discussions went really well; indeed, while NONA offered us a lot in terms of accessibility and sustainability, we found that our team could give NONA a hand as well. |
| + | <br><br> | ||
| + | We agreed to pass Neptune along to the NONA community. Neptune, with its installation link and all of the documentation were passed along to NONA. In the coming weeks more and more of Neptune’s supporting content; wiki’s, tutorials and all documentation will be pushed to NONA’s site. | ||
| + | <br><br> | ||
| + | NONA will now holistically support Neptune and ensure it is a successful, long term open source project. By integrating our software, with all its documentation and supporting educational material, to NONA, we have provided Neptune a community and a platform for long term sustainability. | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | <a href="http://nonasoftware.org/available-software/" style="padding-right:20px"> <button type="button" class="btn btn-primary" id="down">SEE OUR WORK WITH NONA</button></a> | ||
| + | |||
| + | </div> | ||
| + | <div class="col-md-1"></div> | ||
| + | </div> | ||
| + | <div class="row"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
<div class="row"> | <div class="row"> | ||
| − | <div | + | <div class="col-md-4"> |
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | </div> |
| + | <div class="col-md-4"> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/f/fb/T--BostonU_HW--nona_priyak.jpg" width="100%"> | ||
| + | </div> | ||
| + | <div class="col-md-4"> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 665: | Line 695: | ||
</div> | </div> | ||
| + | <br><br><br><br><br><br> | ||
| − | + | <div class="row"> | |
| − | + | ||
| − | + | ||
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> NONA and the iGEM Community </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 678: | Line 707: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | We mentioned that our team could give NONA a hand. Indeed, Neptune is the first iGEM project to be placed on NONA, and in our discussions with NONA we found that our team could pave the way for future iGEM teams that want to have a place for their software tools. | ||
| − | + | <br><br> | |
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| − | |||
| − | <div class="row"> | + | <div class="row"> |
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> The Problem: iGEM software tools get lost </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 708: | Line 726: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | |||
| + | Each year, software solutions to synthetic biology problems are generated from teams around the world. iGEM software tools are especially prone to losing support or community after they are developed, as teams that developed those tools for the competition often find it hard to maintain those projects in the years thereafter. And worse yet, there have been dozens of software tools to come from iGEM, and none of these tools have a clear or coherent locations where they can be found. | ||
| + | |||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| − | |||
<div class="row"> | <div class="row"> | ||
| Line 720: | Line 738: | ||
<div class="row"> | <div class="row"> | ||
<div calss="col-md-4"> | <div calss="col-md-4"> | ||
| − | <img src=" | + | <img src="" width="25%"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 727: | Line 745: | ||
</div> | </div> | ||
| − | + | <br><br> | |
| − | <div class="row"> | + | <div class="row"> |
<div class="col-md-1"> | <div class="col-md-1"> | ||
</div> | </div> | ||
<div class="col-md-11"> | <div class="col-md-11"> | ||
| − | <div style="font-size: 2em; line-height: 150%;"> | + | <div style="font-size: 2em; line-height: 150%;"> The Solution: NONA to protect iGEM software tools </div> |
</div> | </div> | ||
</div> | </div> | ||
| Line 739: | Line 757: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
<div class="col-md-10"> | <div class="col-md-10"> | ||
| − | + | ||
| + | Our team offers itself as a point of contact for NONA at the iGEM Giant Jamboree this year. We want to extend the hand to other iGEM teams that have developed amazing tools, but are worried about how these tools will be maintained in the future. We will have an active NONA “about” booth at our Jamboree table, where we will provide information about what NONA is, how NONA can help iGEM teams with software, and information about how these teams can contribute their tools to NONA. We will also actively help these teams contact NONA and we’ll give them information on how to properly send their tools to NONA, expediting the integration process. | ||
| + | <br><br> | ||
| + | We are very excited that Neptune is now part of the NONA software tool platform. We are even more excited about the incredibly potential of integrating iGEM software tools with NONA. We envision a future where iGEM is integrated with NONA such that NONA has an entire section dedicated to tools developed by iGEM teams. This would be a huge way to make software tools in synthetic biology more integrated, well documented, coherently located and above all, accessible. | ||
| + | |||
</div> | </div> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| − | |||
| − | |||
<div class="row"> | <div class="row"> | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
| Line 751: | Line 771: | ||
<div class="row"> | <div class="row"> | ||
<div calss="col-md-4"> | <div calss="col-md-4"> | ||
| − | <img src=" | + | <img src="" width="25%"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 757: | Line 777: | ||
<div class="col-md-1"></div> | <div class="col-md-1"></div> | ||
</div> | </div> | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | |||
<br><br> | <br><br> | ||
Latest revision as of 03:08, 20 October 2016
HUMAN PRACTICES: GOLD
Neptune: Accessible microfluidics for all
In our Silver Medal Human Practices page, we outlined our public outreach and industry visits. These experiences drew our team to focus on accessibility in synthetic biology as a theme moving forward with our human practice contributions. For Silver HP, we contributed a set of informational blog posts on the history of intellectual property, and IP in software and in synthetic biology today. We welcome you to read these at our WordPress site.
Moving forward with the development of Neptune, we decided that we would extend our silver medal HP theme of accessibility in synthetic biology. Indeed, we were developing a complete, end-to-end microfluidic development workflow. Having seen how inaccessible and prohibitively costly microfluidics are for researchers, and also having studied the virtues of open source tools for synthetic biology, we decided to integrate accessibility into our implementation of Neptune.
In this page, we cover 3 ways in which we expand on and integrate the theme of accessibility to our final product, Neptune.
Moving forward with the development of Neptune, we decided that we would extend our silver medal HP theme of accessibility in synthetic biology. Indeed, we were developing a complete, end-to-end microfluidic development workflow. Having seen how inaccessible and prohibitively costly microfluidics are for researchers, and also having studied the virtues of open source tools for synthetic biology, we decided to integrate accessibility into our implementation of Neptune.
In this page, we cover 3 ways in which we expand on and integrate the theme of accessibility to our final product, Neptune.
- we made it a project criteria that Neptune must interface with low cost, open and readily available tools and hardware to create microfluidics
- we partnered with the NONA Research Foundation, an organization dedicated to increasing access, collaboration, and building a community around synthetic biology software tools
- we offer our team as a point of contact to other iGEM teams that have created software solutions and would like to have these tools protected and stored on NONA
Neptune: Low cost, easy to use, accessible
As we developed Neptune, we were faced with design choices that would have a large impact on how our workflow integrates with the synthetic biology community. In light of our Silver HP discoveries, we decided the most important design choice would be to make our workflow accessible to all: we want a workflow so easy to use that researchers in synthetic biology will be excited to make microfluidic chips. We want a workflow that interfaces with fabrication protocols that are low cost, such that even DIY hobbyists could make new chips. We wanted a workflow whose hardware could be fabricated with a 3D printer, and whose software is open source so that the entire synthetic biology community can contribute to new features in Neptune. Further, we wanted to ensure our workflow completely and cleanly integrates with current fabrication methods, so that we are still following current standards.
Open Source Software
While this is already an iGEM requirement, we are proud to say our source code for Neptune is licensed as open under a BSD-II license. Further, we are very proud to say that Neptune is public on GitHub, and we encourage developers to fork the repo.
Neptune is free to download from GitHub too, and very soon NONA will provide a distribution method.
Neptune is free to download from GitHub too, and very soon NONA will provide a distribution method.
Simple Design Process
Accessibility means ease of use. Ease of use means simplicity; it means a no-brainer user interface and powerful code to help you design microfluidics with ease. Indeed, the biggest setback to designing microfluidics before Neptune was the tedious, long, and difficult process of hand-specifying all features in the microfluidic device. In Neptune, simple english-language descriptions allow you to specify complex microfluidic designs.



Tutorials, Wiki's, Documentation
Not knowing how to use a new tool can dissuade researchers from ever bothering to learn; poor documentation, lack of support for new users, confusing or nonexistent tutorials- all of these add up to a big headache to a researcher who wants to get into microfluidics.
We targeted this issue by creating a tutorial video for our software and hardware workflow. This tutorial is a complete “Hello World” walkthrough that will guide new researchers step-by-step through the microfluidic creation process. We hope that this will encourage anyone who had been intimidated by microfluidic design to come forward and give it a shot.
A tutorial video is good, but not complete. We followed up by starting a extensive written tutorial to writing LFR and MINT files (to design microfluidics,) and written tutorials on how to setup the hardware components. We also added wiki into to the GitHub repositories for Neptune and our hardware, providing yet another resource for new users to try.
We targeted this issue by creating a tutorial video for our software and hardware workflow. This tutorial is a complete “Hello World” walkthrough that will guide new researchers step-by-step through the microfluidic creation process. We hope that this will encourage anyone who had been intimidated by microfluidic design to come forward and give it a shot.
A tutorial video is good, but not complete. We followed up by starting a extensive written tutorial to writing LFR and MINT files (to design microfluidics,) and written tutorials on how to setup the hardware components. We also added wiki into to the GitHub repositories for Neptune and our hardware, providing yet another resource for new users to try.
Integrated, Low Cost Fabrication Protocols
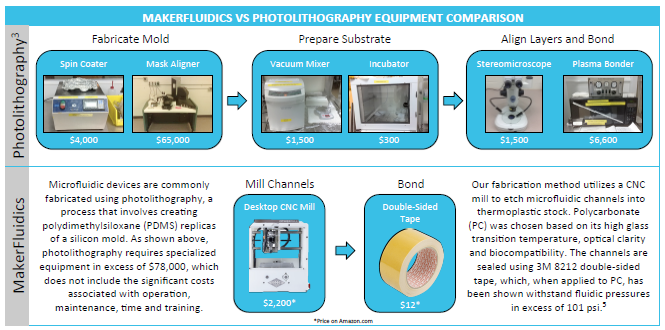
This is truly where the Gold Medal HP contributions become apparent. We had a clear choice here: We could have made Neptune interface with industry grade, powerful and very expensive fabrication methods using photolithography and plasma bonding. If we decided to do this, there would be a blunt eighty-thousand-dollar barrier of entry to researchers and hobbyists looking to use Neptune. That would be unacceptable. We could have had Neptune’s microfluidics be controlled using very high precision, high accuracy pumps that would provide flawless flow rates. But the hardware and infrastructure for these pumps would cost hundreds of dollars per pump, and the cost would not scale. We could not settle for this.
Instead, we framed our workflow around the low cost MakerFluidics fabrication protocol. In this protocol, the eighty thousand dollar fabrication process of photolithography is bypassed; instead our microfluidic chips are fabricated with a simple CNC mill, costing a order of magnitude less, while still delivering high performance devices.
We made it our project goal, to fully integrate accessibility into Neptune. Neptune is microfluidics for industry, for research, for education and for hobbyists. We made Neptune have the lowest possible bar to entry; it is the only microfluidic design, fabrication and control workflow all integrated into one software tool-- and the best part is it can be used by anyone. We made accessibility the centerpiece of Neptune.
Instead, we framed our workflow around the low cost MakerFluidics fabrication protocol. In this protocol, the eighty thousand dollar fabrication process of photolithography is bypassed; instead our microfluidic chips are fabricated with a simple CNC mill, costing a order of magnitude less, while still delivering high performance devices.
We made it our project goal, to fully integrate accessibility into Neptune. Neptune is microfluidics for industry, for research, for education and for hobbyists. We made Neptune have the lowest possible bar to entry; it is the only microfluidic design, fabrication and control workflow all integrated into one software tool-- and the best part is it can be used by anyone. We made accessibility the centerpiece of Neptune.
Open Hardware Schematics
A big aspect of enabling accessible microfluidics is reducing the barrier of entry for controlling these devices. Microfluidics are controlled by pumps that generate pressures gradients to push or pull fluids, or open and close valves. Most modern microfluidic control pumps are expensive and difficult to integrate, making it hard to scale for larger microfluidic designs, and dissuasive for hobbyists.
We chose to directly target our hardware designs to this problem. We developed completely open 3D print design schematics for our microfluidic control infrastructure. These designs are available for download on our GitHub repo, and server though Neptune as well! Furthermore, we developed a completely original hardware design to control syringe pumps using very low cost servo motors.
We chose to directly target our hardware designs to this problem. We developed completely open 3D print design schematics for our microfluidic control infrastructure. These designs are available for download on our GitHub repo, and server though Neptune as well! Furthermore, we developed a completely original hardware design to control syringe pumps using very low cost servo motors.



NONA Research Foundation Partnership
Integrating our workflow with the idea of accessibility was a huge step; we successfully build Neptune as the most accessible microfluidics creation tool to date, with low cost fabrication protocols, open hardware designs, and powerful design algorithms- virtually any research lab can now realistically consider microfluidics as an option, without devoting huge costs or needing expertise in the field.
A big step, but we had to go further. The biggest issue we faced as the summer drew to a close was sustainability. Indeed, a software tool is only accessible if it is properly maintained, updated and if it has a community supporting it. What would happen to Neptune after iGEM? Where will Neptune be in 5 years? If we seriously cared about promoting an accessible workflow, we had to make sure we considered the future of our tool.
A big step, but we had to go further. The biggest issue we faced as the summer drew to a close was sustainability. Indeed, a software tool is only accessible if it is properly maintained, updated and if it has a community supporting it. What would happen to Neptune after iGEM? Where will Neptune be in 5 years? If we seriously cared about promoting an accessible workflow, we had to make sure we considered the future of our tool.
What is NONA?
And so we proceeded to reach out to the NONA Research Foundation, a nonprofit whose aim is to broaden access to and develop a community around open source synthetic biology software tools. As cited from the organization, “Nona creates long-lasting solutions to the challenges posed by the rapidly growing complexity of software in the synthetic biology community by using the power of open source.”
Integrating with NONA provided 3 long-term, sustainable solutions to promoting the accessibility of Neptune and microfluidics to the synthetic biology community.
1) NONA would distribute Neptune, allowing Neptune to have a “home” where is can easily be found (beyond GitHub.)
2) NONA is dedicated to integrating all open synthetic biology software tools under one umbrella, thus building a community of researchers and developers who who will use and support these tools. Thus, NONA will be a medium through which other developers can contribute to Neptune, and NONA will be a place researchers can go to learn more about how to use Neptune, ask for help, and so forth.
3) Overall, by having a dedicated community and website where Neptune will always have a place, we are making it very hard for Neptune to be abandoned. Quite the opposite, with NONA will ensure Neptune is sustained for years to come, while also providing the opportunity for a community to flourish around Neptune and similar software tools.
Integrating with NONA provided 3 long-term, sustainable solutions to promoting the accessibility of Neptune and microfluidics to the synthetic biology community.
1) NONA would distribute Neptune, allowing Neptune to have a “home” where is can easily be found (beyond GitHub.)
2) NONA is dedicated to integrating all open synthetic biology software tools under one umbrella, thus building a community of researchers and developers who who will use and support these tools. Thus, NONA will be a medium through which other developers can contribute to Neptune, and NONA will be a place researchers can go to learn more about how to use Neptune, ask for help, and so forth.
3) Overall, by having a dedicated community and website where Neptune will always have a place, we are making it very hard for Neptune to be abandoned. Quite the opposite, with NONA will ensure Neptune is sustained for years to come, while also providing the opportunity for a community to flourish around Neptune and similar software tools.
Passing Neptune to NONA
We met with members of NONA to discuss integrating Neptune with their foundation. Discussions went really well; indeed, while NONA offered us a lot in terms of accessibility and sustainability, we found that our team could give NONA a hand as well.
We agreed to pass Neptune along to the NONA community. Neptune, with its installation link and all of the documentation were passed along to NONA. In the coming weeks more and more of Neptune’s supporting content; wiki’s, tutorials and all documentation will be pushed to NONA’s site.
NONA will now holistically support Neptune and ensure it is a successful, long term open source project. By integrating our software, with all its documentation and supporting educational material, to NONA, we have provided Neptune a community and a platform for long term sustainability.
We agreed to pass Neptune along to the NONA community. Neptune, with its installation link and all of the documentation were passed along to NONA. In the coming weeks more and more of Neptune’s supporting content; wiki’s, tutorials and all documentation will be pushed to NONA’s site.
NONA will now holistically support Neptune and ensure it is a successful, long term open source project. By integrating our software, with all its documentation and supporting educational material, to NONA, we have provided Neptune a community and a platform for long term sustainability.

NONA and the iGEM Community
We mentioned that our team could give NONA a hand. Indeed, Neptune is the first iGEM project to be placed on NONA, and in our discussions with NONA we found that our team could pave the way for future iGEM teams that want to have a place for their software tools.
The Problem: iGEM software tools get lost
Each year, software solutions to synthetic biology problems are generated from teams around the world. iGEM software tools are especially prone to losing support or community after they are developed, as teams that developed those tools for the competition often find it hard to maintain those projects in the years thereafter. And worse yet, there have been dozens of software tools to come from iGEM, and none of these tools have a clear or coherent locations where they can be found.
The Solution: NONA to protect iGEM software tools
Our team offers itself as a point of contact for NONA at the iGEM Giant Jamboree this year. We want to extend the hand to other iGEM teams that have developed amazing tools, but are worried about how these tools will be maintained in the future. We will have an active NONA “about” booth at our Jamboree table, where we will provide information about what NONA is, how NONA can help iGEM teams with software, and information about how these teams can contribute their tools to NONA. We will also actively help these teams contact NONA and we’ll give them information on how to properly send their tools to NONA, expediting the integration process.
We are very excited that Neptune is now part of the NONA software tool platform. We are even more excited about the incredibly potential of integrating iGEM software tools with NONA. We envision a future where iGEM is integrated with NONA such that NONA has an entire section dedicated to tools developed by iGEM teams. This would be a huge way to make software tools in synthetic biology more integrated, well documented, coherently located and above all, accessible.
We are very excited that Neptune is now part of the NONA software tool platform. We are even more excited about the incredibly potential of integrating iGEM software tools with NONA. We envision a future where iGEM is integrated with NONA such that NONA has an entire section dedicated to tools developed by iGEM teams. This would be a huge way to make software tools in synthetic biology more integrated, well documented, coherently located and above all, accessible.

