(→Effect of farnesol on the growth of Staphylococcus aureus) |
|||
| (17 intermediate revisions by 3 users not shown) | |||
| Line 585: | Line 585: | ||
=='''Effect of farnesol on the growth of Staphylococcus aureus'''== | =='''Effect of farnesol on the growth of Staphylococcus aureus'''== | ||
We examined whether farnesol also has an effect against food poisoning bacteria. We used Staphylococcus aureus as food poisoning bacteria. We examined whether farnesol inhibits their growth. | We examined whether farnesol also has an effect against food poisoning bacteria. We used Staphylococcus aureus as food poisoning bacteria. We examined whether farnesol inhibits their growth. | ||
| − | [[File:阻止円実験.jpg| | + | [[File:阻止円実験.jpg|800px|thumb|center|State of the experiment:Effect of fragrance for growth of microbial]] |
<br /> | <br /> | ||
[[File:result_soshien ousyokubudoukyukin.jpg|800px|thumb|center|Fig5:Effect of farnesol on the growth of Staphylococcus aureus | [[File:result_soshien ousyokubudoukyukin.jpg|800px|thumb|center|Fig5:Effect of farnesol on the growth of Staphylococcus aureus | ||
| Line 610: | Line 610: | ||
While, inhibition circle was observed on the plate with farnesol (A).]] | While, inhibition circle was observed on the plate with farnesol (A).]] | ||
<br /> | <br /> | ||
| − | This result indicates that farnesol affect ''E.coli''. So, we | + | This result indicates that farnesol affect ''E.coli''. So, we hypothesized that ''E.coli'' needs to have resistance to farnesol. |
<br /> | <br /> | ||
| Line 623: | Line 623: | ||
=Confirmation of Amount of transcription of Farnesol production device.= | =Confirmation of Amount of transcription of Farnesol production device.= | ||
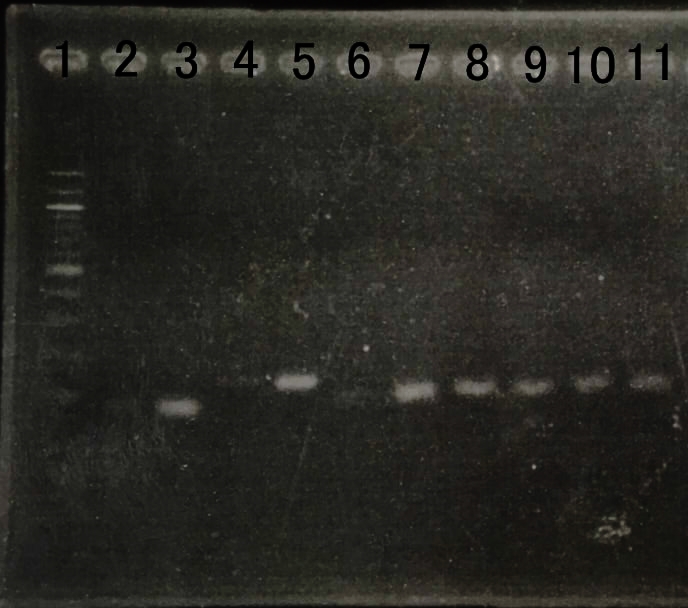
| + | We reinforced by overexpressing genes that dxs,idi,ispD and ispF, which was the bottom neck of MEP pathway. | ||
| + | We checked amount of transcription by RT-PCR. | ||
| + | E. coli that we created last year can overexpress the gene. | ||
| + | We checked amount of transcription by RT-PCR to confirm whether E.coli can overexpress the gene or not. | ||
| + | We thought that amount of RNA transcription was increased . | ||
| − | + | [[File:RT-PCR MEP.jpg|800px|thumb|center|Fig8.The result of RT-PCR | |
| + | <br> | ||
| + | Lane1:100bp DNA ladder | ||
| + | <br> | ||
| + | Lane2:dxs(WT) | ||
| + | <br> | ||
| + | Lane3:dxs(recombinant) | ||
| + | <br> | ||
| + | Lane4:idi(WT) | ||
| + | <br> | ||
| + | Lane5:idi(recombinant) | ||
| + | <br> | ||
| + | Lane6:ispA(WT) | ||
| + | <br> | ||
| + | Lane7:ispA(recombinant) | ||
| + | <br> | ||
| + | Lane8:ispD(WT) | ||
| + | <br> | ||
| + | Lane9:ispD(recombinant) | ||
| + | <br> | ||
| + | Lane10:gapA(WT) | ||
| + | <br> | ||
| + | Lane12:gapA(recombinant) ]] | ||
| − | + | It is a possibility that cDNA is mixed to gDNA . | |
| − | [[File:RT-PCR MEP.jpg|800px|thumb|center| | + | In order to investigate whether gDNA was remove or not we measured gDNA as a negative control by RT-PCR. |
| + | |||
| + | |||
| + | <br> | ||
| + | [[File:RT-PCR MEP Negative control.jpg|800px|thumb|center|Fig9.The result of RT-PCR Negative control | ||
<br> | <br> | ||
Lane1:100bp DNA ladder | Lane1:100bp DNA ladder | ||
| + | <br> | ||
Lane2:dxs(WT) | Lane2:dxs(WT) | ||
| + | <br> | ||
Lane3:dxs(recombinant) | Lane3:dxs(recombinant) | ||
| + | <br> | ||
Lane4:idi(WT) | Lane4:idi(WT) | ||
| + | <br> | ||
Lane5:idi(recombinant) | Lane5:idi(recombinant) | ||
| + | <br> | ||
Lane6:ispA(WT) | Lane6:ispA(WT) | ||
| + | <br> | ||
Lane7:ispA(recombinant) | Lane7:ispA(recombinant) | ||
| + | <br> | ||
Lane8:ispD(WT) | Lane8:ispD(WT) | ||
| + | <br> | ||
Lane9:ispD(recombinant) | Lane9:ispD(recombinant) | ||
| + | <br> | ||
Lane10:gapA(WT) | Lane10:gapA(WT) | ||
| + | <br> | ||
Lane12:gapA(recombinant) ]] | Lane12:gapA(recombinant) ]] | ||
| + | =Confirmation of Amount of transcription of PgpB.device, YbjG.device, and IspC-ribB(G108S).device= | ||
| + | We thought that gDNA could be removed ,because band was not appeared any lane. | ||
| + | |||
| + | Therefore, we thought that the amount of transcription of pgpB was increased. | ||
| + | [[File:RT-PCR ybjG N.C..jpg|800px|thumb|center|Fig10.The result of RT-PCR ybjG pgpB Fusion | ||
| + | <br> | ||
| + | Lane1:100bp DNA ladder | ||
| + | <br> | ||
| + | Lane2:ispC(WT) | ||
| + | <br> | ||
| + | Lane3:ispC(Fusion) | ||
| + | <br> | ||
| + | Lane4:ybjG (WT) | ||
| + | <br> | ||
| + | Lane5:ybjG (ybjG) | ||
| + | <br> | ||
| + | Lane6:pgpB (WT) | ||
| + | <br> | ||
| + | Lane7:pgpB (pgpB) | ||
| + | <br> | ||
| + | Lane8:gapA(WT) | ||
| + | <br> | ||
| + | Lane9:gapA (Fusion) | ||
| + | <br> | ||
| + | Lane10:gapA (ybjG) | ||
| + | <br> | ||
| + | Lane11:gapA (pgpB)]] | ||
| + | |||
| + | It is a possibility that cDNA is mixed to gDNA . | ||
| + | |||
| + | In order to investigate whether gDNA was remove or not we measured gDNA as a negative control by RT-PCR. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | [[File:Photo.jpg|800px|thumb|center|Fig10.The result of RT-PCR ybjG pgpB Fusion negative control | ||
| + | <br> | ||
| + | Lane1:100bp DNA ladder | ||
| + | <br> | ||
| + | Lane2:ispC(WT) | ||
| + | <br> | ||
| + | Lane3:ispC(Fusion) | ||
| + | <br> | ||
| + | Lane4:ybjG (WT) | ||
| + | <br> | ||
| + | Lane5:ybjG (ybjG) | ||
| + | <br> | ||
| + | Lane6:pgpB (WT) | ||
| + | <br> | ||
| + | Lane7:pgpB (pgpB) | ||
| + | <br> | ||
| + | Lane8:gapA(WT) | ||
| + | <br> | ||
| + | Lane9:gapA (Fusion) | ||
| + | <br> | ||
| + | Lane10:gapA (ybjG) | ||
| + | <br> | ||
| + | Lane11:gapA (pgpB)]] | ||
| + | |||
| + | We thought that gDNA could be removed ,because band was not appeared any lane. | ||
| + | Therefore, we thought that the amount of transcription of pgpB was increased. | ||
| + | <br> | ||
| + | Whether the fusion protein is actually connected it was confirmed by measuring the amount of transcription at ispC and mriB. | ||
| + | To measure the amount of transcription ispC for the gene of the fusion protein to confirm whether they are transferred | ||
[[File:RT-PCR fusion.jpg|800px|thumb|center|Fig2.The result of RT-PCR fusion protein | [[File:RT-PCR fusion.jpg|800px|thumb|center|Fig2.The result of RT-PCR fusion protein | ||
<br> | <br> | ||
Lane1:100bp DNA ladder | Lane1:100bp DNA ladder | ||
| + | <br> | ||
Lane2:ispC(WT) | Lane2:ispC(WT) | ||
| + | <br> | ||
Lane3:ispC(recombinant) | Lane3:ispC(recombinant) | ||
| + | <br> | ||
Lane4:m-ribB(WT) | Lane4:m-ribB(WT) | ||
| + | <br> | ||
Lane5:m-ribB(recombinant) | Lane5:m-ribB(recombinant) | ||
| + | <br> | ||
Lane6:gapA(WT) | Lane6:gapA(WT) | ||
| + | <br> | ||
Lane7:gapA(recombinant) | Lane7:gapA(recombinant) | ||
| + | <br> | ||
Lane8:ispC(WT) Negative control | Lane8:ispC(WT) Negative control | ||
| + | <br> | ||
Lane9:ispC(recombinant) Negative control | Lane9:ispC(recombinant) Negative control | ||
| + | <br> | ||
Lane10:m-ribB(WT) Negative control | Lane10:m-ribB(WT) Negative control | ||
| + | <br> | ||
Lane11:m-ribB(recombinant)Negative control | Lane11:m-ribB(recombinant)Negative control | ||
| + | <br> | ||
Lane12:gapA(WT) Negative control | Lane12:gapA(WT) Negative control | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | + | Lane13:gapA(recombinant)Negative control]] | |
| − | + | From this result, the transcription of mribB and ispC is being performed, it can be said that the construct of the fusion protein was successful | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 03:51, 20 October 2016
Contents
- 1 Confirmation antibacterial activity of farnesol.
- 2 Confirmation of anti-mold, anti-maggots and antibacterial activities of farnesol.
- 3 Enhancement of farnesol resistance
- 4 Confirmation of Amount of transcription of Farnesol production device.
- 5 Confirmation of Amount of transcription of PgpB.device, YbjG.device, and IspC-ribB(G108S).device
Confirmation antibacterial activity of farnesol.
First, we examined our working hypothesis to “Flavorator” that farnesol can show either the antibacterial or bacteriostatic activity in a box like “KOZOKO”. The results clearly showed that farnesol had antibacterial properties. In the literatures, farnesol have antibacterial volatiles. Farnesol is produced after complicated pathways, so for their syntheses, various enzymes are required. In E.coli, farnesol may be synthesized. In this context, we designed our system for establishing the concept of “Flavorator” to build up a brand-new biosynthetic pathways, in which farnesol is produced in the E.coli. In doing so, we transfered the three types of genes listed below to create the hyper-producer E.coli of farnesol. We examined our working hypothesis to “Flavorator” that farnesol can show either the antibacterial or bacteriostatic activity in a box like Kozoko.
Confirmation of anti-mold, anti-maggots and antibacterial activities of farnesol.
E.coli can easily synthesize antimicrobial volatiles farnesol and geraniol. We examined the effects of three antimicrobial volatiles against bacteria which rot food.
Comparison of the suppressing effect of geraniol, farnesol and ethanol on mold

A: geraniol(100 μl of geraniol solution was dropped on the cotton)B:farnesol(100 μl) C: ethanol (100 μl ) A:Chopstick dipped in suspension of mold was touched in the center of the bread. The volume of box is 846cm³. B:The same treatment was done. C:The same treatment was done. These boxes were kept for 5 days at room temperature.
From this result, farnesol proved that it has the best anti-fungal activity against mold.
farnesol has a preservative effect on various foods
Farnesol has high antifungal activity against the mold of bread. Therefore, we investigated whether farnesol exerts similar antifungal effects on other food.
Effects of farnesol on bread mold.

A: farnesol(1ml of farnesol solution was dropped on the cotton), B:ddH2O(1ml). A:Chopstick dipped in suspension of mold was touched in the center of the bread. The volume of box is 846cm³. B: The same treatment was done. These boxes were kept for 8 days at room temperature.
Effects of farnesol on rice mold.

A: farnesol(1ml of farnesol solution was dropped on the cotton), B:ddH2O(1ml). A:Chopstick dipped in suspension of mold was touched in the center of the rice. The volume of box is 846cm³. B: The same treatment was done. These boxes were kept for 8 days at room temperature.
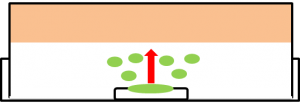
Effects of farnesol on maggot.
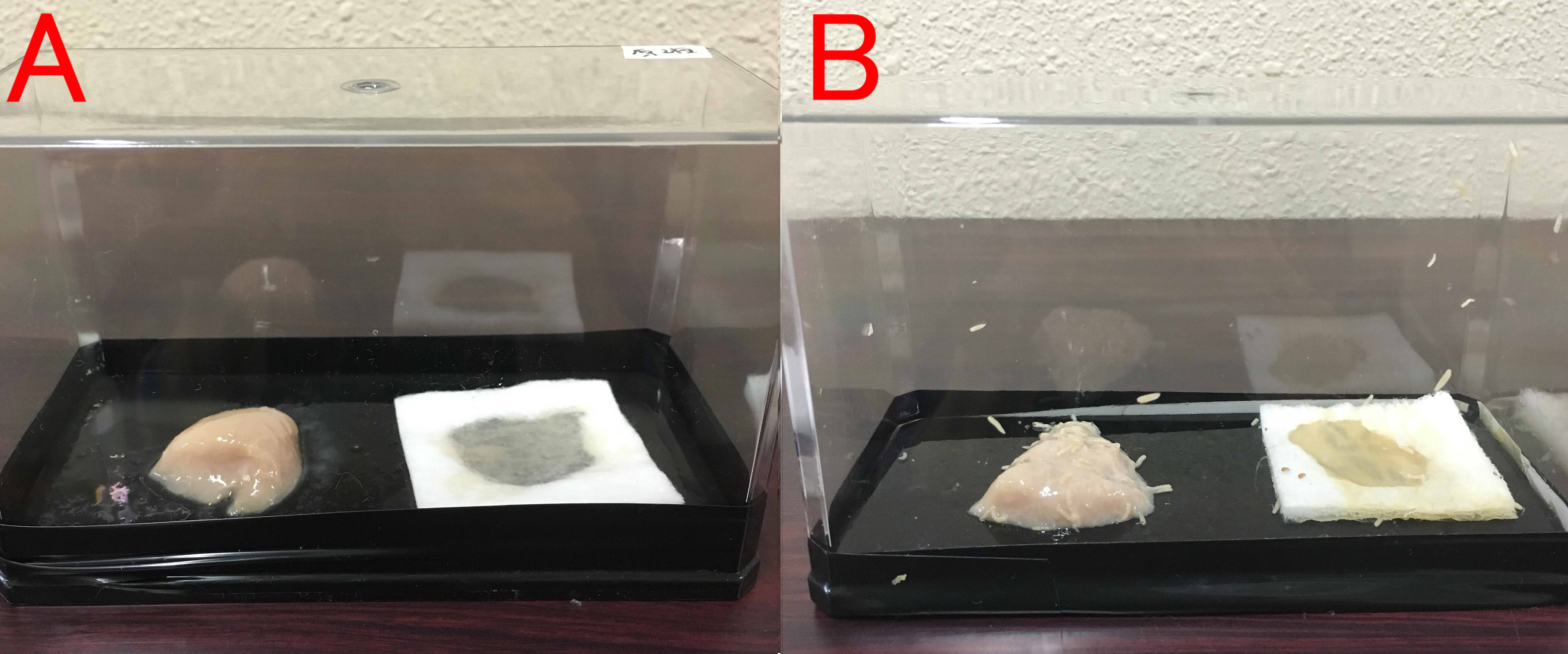
A: farnesol(1ml of farnesol solution was dropped on the cotton), B:ddH2O(1ml). A:Chopstick dipped in suspension of rotten meat was touched in the center of the chicken. The volume of box is 846cm³. B: The same treatment was done. These boxes were kept for 8 days at room temperature.
We found that farnesol has a preservative effect on various foods.
Effect of farnesol on the growth of Staphylococcus aureus
We examined whether farnesol also has an effect against food poisoning bacteria. We used Staphylococcus aureus as food poisoning bacteria. We examined whether farnesol inhibits their growth.
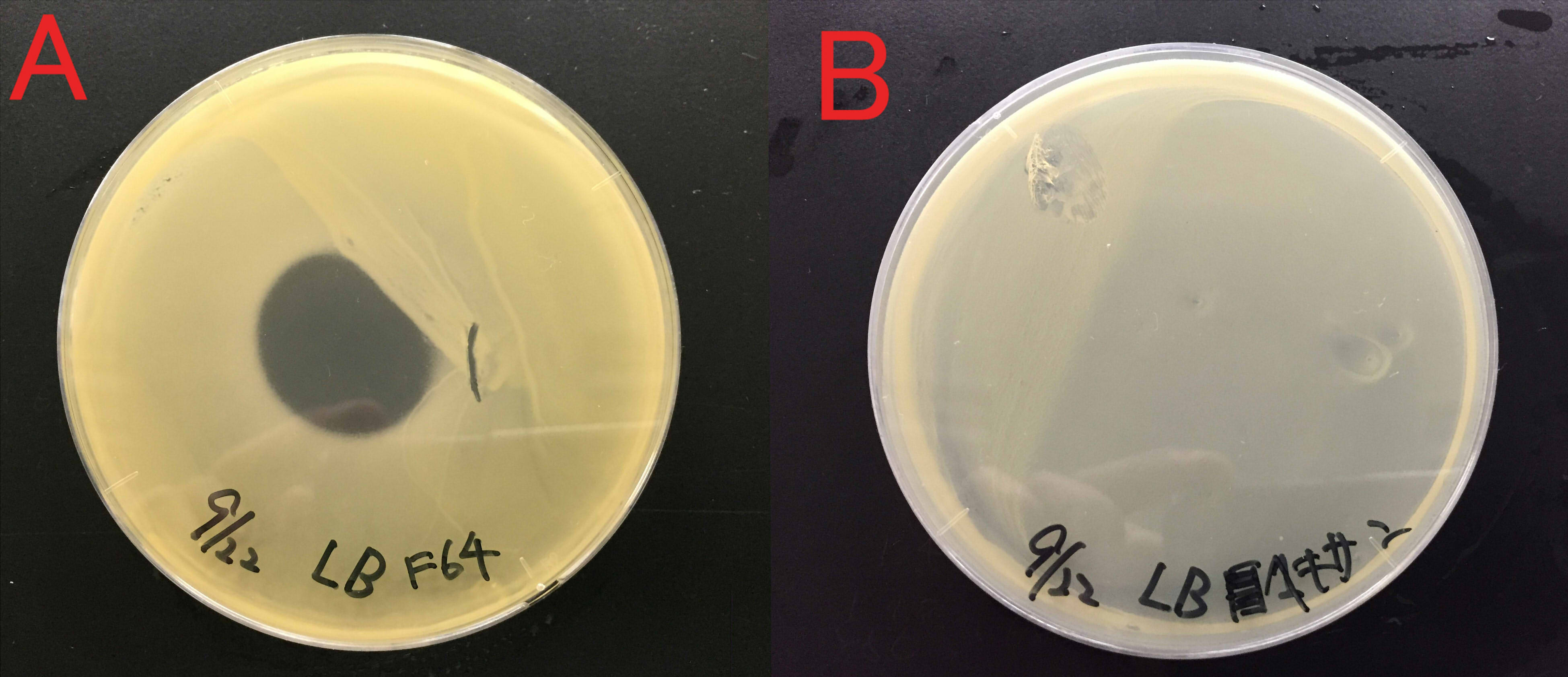
A:Farnesol with ddH2O(300μl) B:ddH2O(300μl). Farnesol was dropped on the paper which was put on center of the plate cover without direct contact with the bacteria. These plates were incubated for 21 hours at 37 ℃. Growth inhibition circle was not observed on the plate without farnesol (B). While, inhibition circle was observed on the plate with farnesol (A). These results indicate that it is difficult for Staphylococcus aureus to grow in the presence of farnesol.
We considered that farnesol has high effect against inhibitory the growth food poisoning bacteria because diluted farnesol affect staphylococcus aureus.
Effect of farnesol on growth of E.coli
We thought about a possibility that farnesol may affect E.coli, which produces farnesol, because E.coli is also a bacterium. Therefore, we examined whether farnesol affects E.coli.
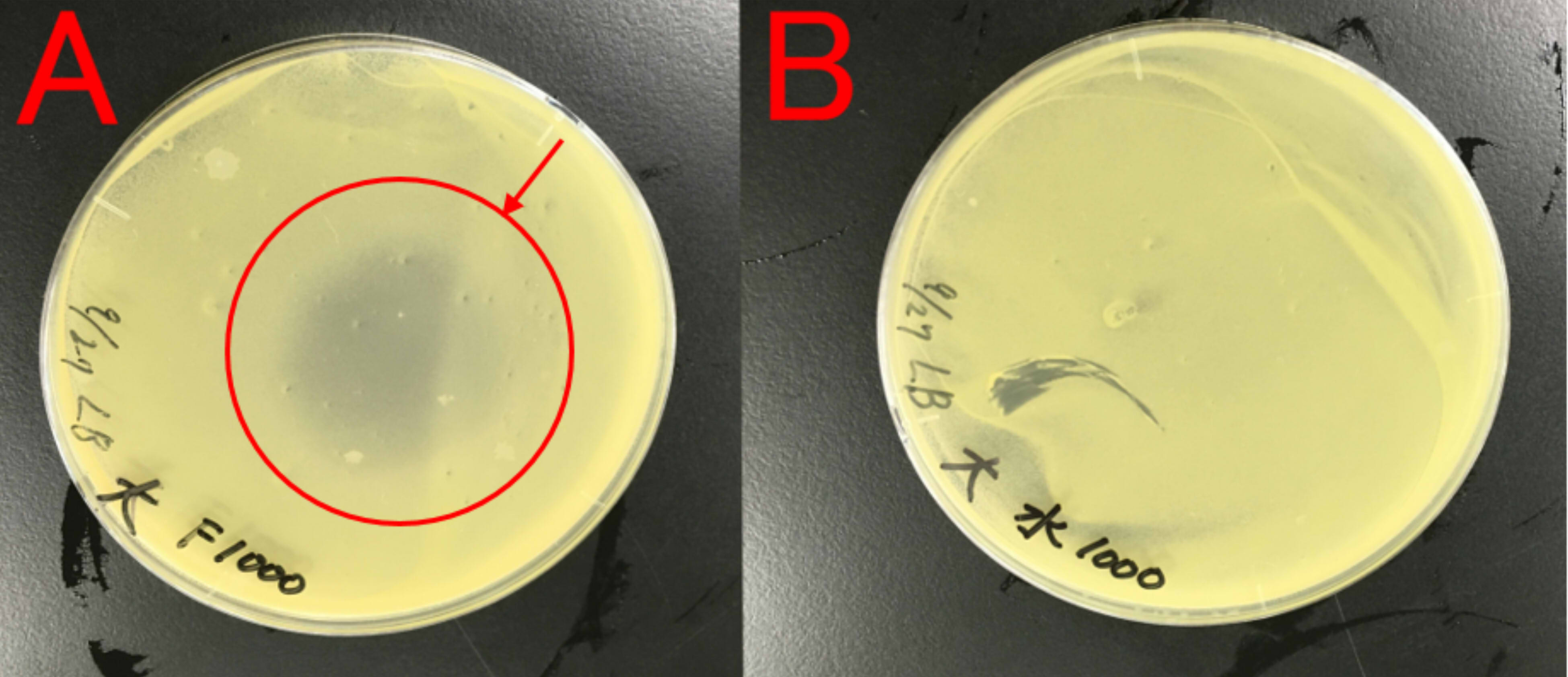
A: farnesol (1ml) B: ddH2O (1ml) Farnesol was dropped on the paper which was put on center of the plate cover without direct contact with the bacteria. These plates were incubated for 21 hours at 37 ℃. Growth inhibition circle was not observed on the plate(B). While, inhibition circle was observed on the plate with farnesol (A).
This result indicates that farnesol affect E.coli. So, we hypothesized that E.coli needs to have resistance to farnesol.
Enhancement of farnesol resistance
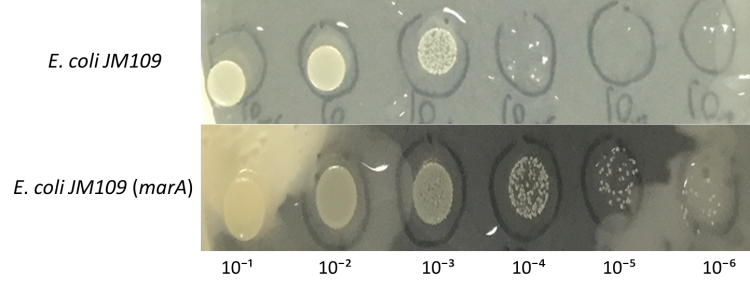
E.coli JM109 and E.coli JM109 (marA) were dropped on LBGMg agar plates in serial ten-fold dilutions (10⁻¹~〖10〗^(-6)), overlaid with 30 % (v/v) farnesol hexane solution (farnesol solution) and incubated at 30°C for 24 h. This figure shows that E.coli JM109 (marA) cells that overexpress the marA product better than the control E.coli JM109 wild type cells survived plates overloyed by 30 % farnesol solution.
Confirmation of Amount of transcription of Farnesol production device.
We reinforced by overexpressing genes that dxs,idi,ispD and ispF, which was the bottom neck of MEP pathway. We checked amount of transcription by RT-PCR. E. coli that we created last year can overexpress the gene. We checked amount of transcription by RT-PCR to confirm whether E.coli can overexpress the gene or not. We thought that amount of RNA transcription was increased .
It is a possibility that cDNA is mixed to gDNA .
In order to investigate whether gDNA was remove or not we measured gDNA as a negative control by RT-PCR.
Confirmation of Amount of transcription of PgpB.device, YbjG.device, and IspC-ribB(G108S).device
We thought that gDNA could be removed ,because band was not appeared any lane.
Therefore, we thought that the amount of transcription of pgpB was increased.
It is a possibility that cDNA is mixed to gDNA .
In order to investigate whether gDNA was remove or not we measured gDNA as a negative control by RT-PCR.
We thought that gDNA could be removed ,because band was not appeared any lane.
Therefore, we thought that the amount of transcription of pgpB was increased.
Whether the fusion protein is actually connected it was confirmed by measuring the amount of transcription at ispC and mriB.
To measure the amount of transcription ispC for the gene of the fusion protein to confirm whether they are transferred
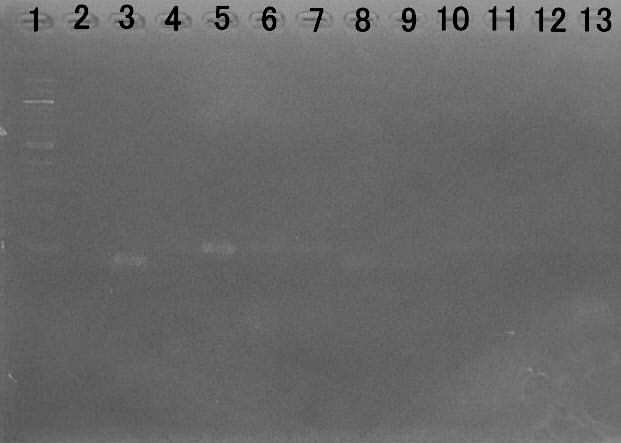
Lane1:100bp DNA ladder
Lane2:ispC(WT)
Lane3:ispC(recombinant)
Lane4:m-ribB(WT)
Lane5:m-ribB(recombinant)
Lane6:gapA(WT)
Lane7:gapA(recombinant)
Lane8:ispC(WT) Negative control
Lane9:ispC(recombinant) Negative control
Lane10:m-ribB(WT) Negative control
Lane11:m-ribB(recombinant)Negative control
Lane12:gapA(WT) Negative control
Lane13:gapA(recombinant)Negative control
From this result, the transcription of mribB and ispC is being performed, it can be said that the construct of the fusion protein was successful