| (32 intermediate revisions by 3 users not shown) | |||
| Line 136: | Line 136: | ||
<!-- theme --> | <!-- theme --> | ||
<!-- word begin--> | <!-- word begin--> | ||
| − | + | <div> <b><span style="font-size:11pt;font-family:Arial, sans-serif">Basic Part</span></b><br/> | |
| − | <div> | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">Cry4Aa4+Extended FMDV 2A-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Cry4Aa4+Extended FMDV 2A</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in DH5α</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">3673bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
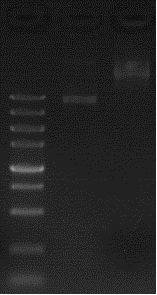
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs1.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs2.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">Cry10Aa4+Extended FMDV 2A-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in Top10</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2158bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs3.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs4.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">Cry11Aa4+Extended FMDV 2A-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Cry11Aa4+ Extended FMDV 2A</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in JM108</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2071bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs3.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs6.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><i><span style="font-size:9.0pt;font-family:":,sans-serif;color:#595959"> </span></i><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | |||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">Cyt1-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Cyt1</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in JM108</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">762bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs3.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs8.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><i><span style="font-size:9.0pt;font-family:":,sans-serif;color:#595959"> </span></i><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">Cyt2-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Cyt1</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in JM108</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">801bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td colspan="2" valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs3.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs10.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td colspan="2" valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td width="54" valign="top" style="width:40.5pt;padding:0cm 0cm 0cm 0cm"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Tahoma, sans-serif"> </span></p> | ||
| + | </td> </tr> <tr> <td width="138" valign="top" style="width:103.5pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="45" valign="top" style="width:33.75pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="124" valign="top" style="width:93.0pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="107" valign="top" style="width:80.25pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="80" valign="top" style="width:60.0pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="2" valign="top" style="width:1.5pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> <td width="54" valign="top" style="width:40.5pt;padding:7.5pt 7.5pt 7.5pt 7.5pt"></td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | = <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79">EGFP-pChlamy_3</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> = | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Name</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">06-EGFP</span><span style="font-size:11pt;font-family:宋体">(</span><span style="font-size:11pt;font-family:Calibri, sans-serif">TAA</span><span style="font-size:11pt;font-family:宋体">)</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Vector</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">pChlamy_3</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Construct Resistant</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Ampicillin in Top10</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Gene Length</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">732bp</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Cloning Strategy</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">QC Items</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Method</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Specifications</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><b><span style="font-size:12pt;font-family:Calibri, sans-serif">Results</span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Target Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequencing results are consistent to the confirmed sequence.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Vector Sequence</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">20bp flanking sequences of the vector are correct.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Reading Frame</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Sequence alignment</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Frame is correct and consistent to the client's requirement.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Fragment Size</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Restriction Digests</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">The size of inserted fragment is right and free of any contaminated bands.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Quality/Quantity</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">UV spectrophotometry</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Miniprep: OD260/280=1.7~2.0</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">2µg/tube, 1 tube,Lyophilized</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Type of Strain Stock</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Bacteria stock.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-top:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Appearance</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Visual inspection</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif">Clear and free of foreign particles, and the strain is clearly visible.</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border-top:none;border-left:none;border-bottom:solid #A3A3A3 1.0pt;border-right:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="center" style="margin-top:4.8pt;margin-right:0cm;margin-bottom:6.0pt;margin-left:0cm;text-align:center"><span style="font-size:11pt;font-family:Calibri, sans-serif">Pass</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | |||
| + | <table border="0" cellspacing="0" cellpadding="0" width="823" style="width:617.25pt;border-collapse:collapse"> | ||
| + | <tr> <td valign="top" style="border:solid #A3A3A3 1.0pt;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs3.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:12pt;font-family:宋体"> [[File:T--FAFU-CHINA--bs12.png | 825px | thumb | center ]]</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> <td valign="top" style="border:solid #A3A3A3 1.0pt;border-left:none;padding:2.0pt 3.0pt 2.0pt 3.0pt"> <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane M: DNA Marker</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 1: Plasmid digested by XbaI and PstI</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:10pt;font-family:Calibri, sans-serif">Lane 2: Plasmid DNA</span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><span style="font-size:11pt;font-family:Calibri, sans-serif"> </span><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
| + | </td> </tr> | ||
| + | </table> | ||
| + | <p align="left" style="margin:4.8pt 0cm 6pt"><b><span style="font-size:16.0pt;font-family:":,sans-serif;color:#1E4E79"> </span></b><span style="font-size:10pt;font-family:Tahoma, sans-serif"></span></p> | ||
<!-- word end --> | <!-- word end --> | ||
| Line 1,547: | Line 563: | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<script> | <script> | ||
$('#page-contents a').click(function() { | $('#page-contents a').click(function() { | ||
Latest revision as of 06:36, 8 November 2016
Contents
Cry4Aa4+Extended FMDV 2A-pChlamy_3
| Gene Name | Cry4Aa4+Extended FMDV 2A |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in DH5α |
| Gene Length | 3673bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results |
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass |
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass |
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass |
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass |
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass |
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass |
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass |
|
|
| Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA |
Cry10Aa4+Extended FMDV 2A-pChlamy_3
| Gene Name | |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in Top10 |
| Gene Length | 2158bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results |
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass |
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass |
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass |
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass |
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass |
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass |
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass |
|
|
| Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA |
Cry11Aa4+Extended FMDV 2A-pChlamy_3
| Gene Name | Cry11Aa4+ Extended FMDV 2A |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in JM108 |
| Gene Length | 2071bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results |
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass |
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass |
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass |
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass |
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass |
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass |
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass |
|
|
| Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA
|
Cyt1-pChlamy_3
| Gene Name | Cyt1 |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in JM108 |
| Gene Length | 762bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results |
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass |
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass |
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass |
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass |
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass |
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass |
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass |
|
|
|
Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA
|
Cyt2-pChlamy_3
| Gene Name | Cyt1 |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in JM108 |
| Gene Length | 801bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results | |||
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass | |||
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass | |||
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass | |||
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass | |||
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass | |||
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass | |||
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass | |||
|
|
|
Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA
|
| |||
EGFP-pChlamy_3
| Gene Name | 06-EGFP(TAA) |
| Cloning Vector | pChlamy_3 |
| Construct Resistant | Ampicillin in Top10 |
| Gene Length | 732bp |
| Cloning Strategy | XbaI and PstI |
| QC Items | Method | Specifications | Results |
| Target Sequence | Sequence alignment | Sequencing results are consistent to the confirmed sequence. | Pass |
| Vector Sequence | Sequence alignment | 20bp flanking sequences of the vector are correct. | Pass |
| Reading Frame | Sequence alignment | Frame is correct and consistent to the client's requirement. | Pass |
| Fragment Size | Restriction Digests | The size of inserted fragment is right and free of any contaminated bands. | Pass |
| Quality/Quantity | UV spectrophotometry | Miniprep: OD260/280=1.7~2.0 2µg/tube, 1 tube,Lyophilized | Pass |
| Type of Strain Stock | Visual inspection | Bacteria stock. | Pass |
| Appearance | Visual inspection | Clear and free of foreign particles, and the strain is clearly visible. | Pass |
|
|
|
Lane M: DNA Marker
Lane 1: Plasmid digested by XbaI and PstI
Lane 2: Plasmid DNA
|