| Line 214: | Line 214: | ||
<a href="https://2016.igem.org/Team:BostonU_HW/HP/Gold">Gold</a> | <a href="https://2016.igem.org/Team:BostonU_HW/HP/Gold">Gold</a> | ||
| − | |||
</div> | </div> | ||
</li> | </li> | ||
Latest revision as of 03:10, 20 October 2016
COLLABORATIONS
NEPTUNE Collaborative Findings
Given that Neptune was built to provide an accessible, affordable, and convenient solution for designing, manufacturing, and operating microfluidic systems, it was only natural that we would test our toolchain by designing microfluidics for other iGEM wetlab teams. We were able to collaborate with both the MIT iGEM team and the Northeastern iGEM team. Through these exchanges, we were able to test the Neptune workflow while providing a wetlab team with microfluidics to test their biological systems.
Designing Automated Mammalian Cell Culture Devices
SEPT - OCT 2016 | A collaboration with the MIT 2016 iGEM Wetlab Team
Our collaboration with the MIT iGEM team involved designing, creating, and controlling microfluidic devices to culture mammalian cells on chip. The end goal of these experiments was to lay the groundwork for testing endometrial cell lines and their response to various levels of estrogen in an automated microfluidic setting.
PART 1
In our first on-site visit, we designed and fabricated a microfluidic device with specific parameters fit to trapping the MIT team's mammalian HEK293 cells. Our first iteration of chip included an input, cell traps surrounded by two valves, and an output, as shown. We automated the control of the valves using our servo-syringe combination pumps. To prepare the chip for the cells, first, we opened our valves to flow 1.2mL of ethanol solution inside the chip for sterilization. Next, we pulled 1mL of PBS (buffer solution) to clean out the ethanol solution for the benefit of the cells. Finally, we pulled 1ml of cell solution with a concentration of 1 x 10^6 cells/ml through the microfluidic chip. The two valves were then closed (trapping the cells and culture media inside) and the chip removed from the control infrastructure. From there, we observed masses of cells moving in the sealed area of the chip under a standard light microscope at 10x, as shown. Finally, we relocated the chip an incubator to grow the cells. Unfortunately, the device dried out a day after being placed in the incubator. This visit proved our fabrication flow's ability to create microfluidic devices capable of trapping HEK293 mammalian cells, and gave both teams great insight on the other end of the biologist-microfluidics bridge, and sparked ideas for further customization of microfluidics for the lab bench. Highlighting the rapid design and prototyping enabled with Neptune, not a week later we returned with an entirely new design and batch of two chips ready for further testing (while our design and fabrication flow allows for iterations to occur within hours, the schedule of both teams needed about a week). This further collaboration is detailed below.
{Attributions/Disclaimer: Present were Shane McCormack, Priya Kapadia, and mentor Radhakrishna Sanka of the BU iGEM HW team, as well as Wangui Mbuguiro and mentor Brian Teague of the MIT iGEM team. All handling of biological materials performed by Wangui Mbuguiro of the MIT iGEM team}
{Attributions/Disclaimer: Present were Shane McCormack, Priya Kapadia, and mentor Radhakrishna Sanka of the BU iGEM HW team, as well as Wangui Mbuguiro and mentor Brian Teague of the MIT iGEM team. All handling of biological materials performed by Wangui Mbuguiro of the MIT iGEM team}

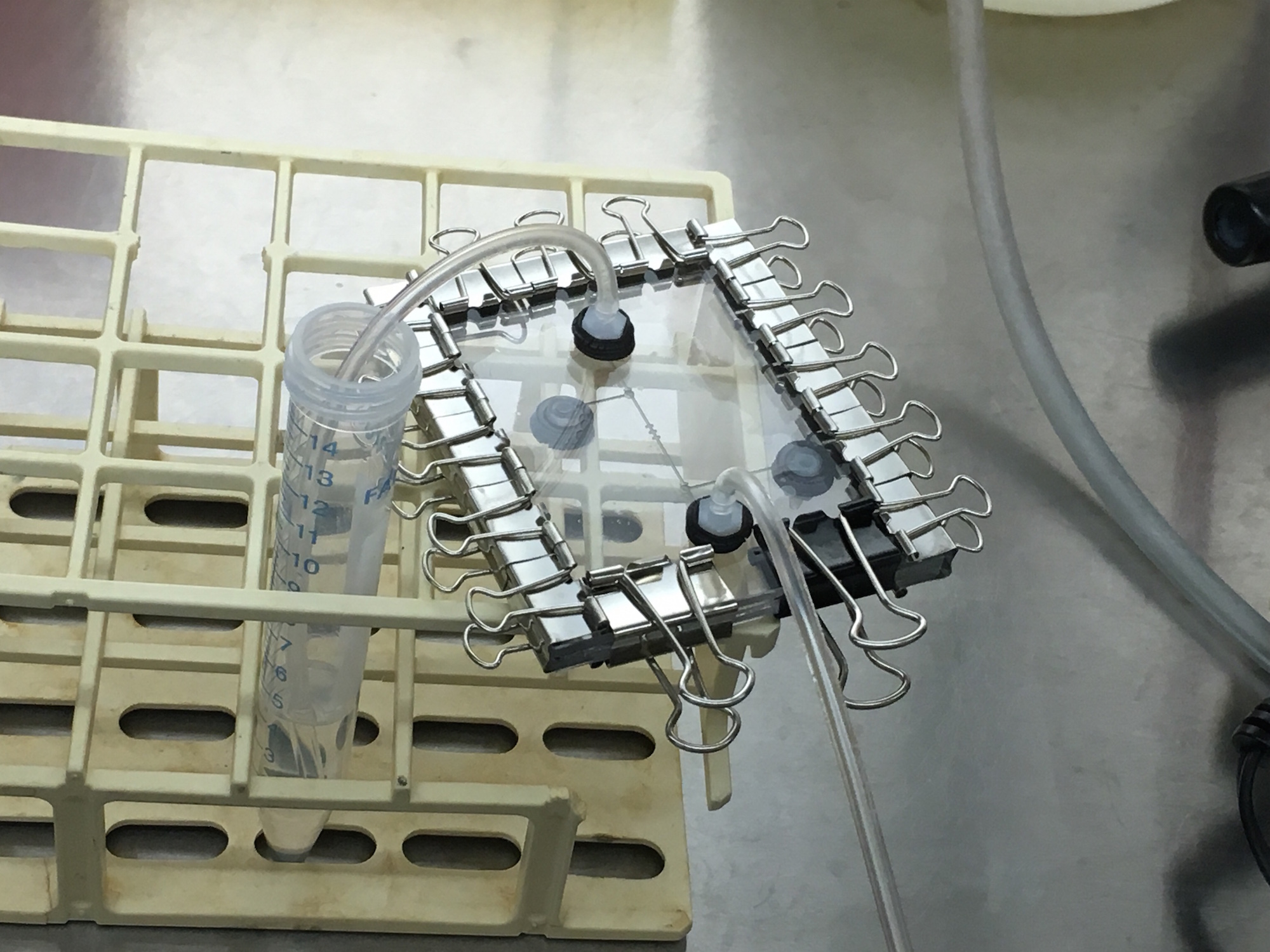



PART 2
For our second on-site visit to MIT, we redesigned our chip to better observe the HEK293 cells under the microscope. Namely, this iteration replaced the cell traps and valves with a diamond chamber and an equivalently shaped window through the polycarbonate for facilitated microscopy on the otherwise empty control (red) layer. The valves were removed with the hopes of running fluid through the device while it was under the microscope, and not detaching it from its pumping infrastructure like in the previous iteration. The fluids pumped through the device were similar to the first iteration, starting with ethanol solution, then PBS, suspended cells mixed with culture media, and finally culture media alone. Evaporation, which occurred in the previous chip in the incubator, was assumed to not be in effect in the closed system of the microscope. To allow the cells time to adhere to the PDMS, the flow of culture media was disrupted; unfortunately the flow was disrupted for too long, and the fluid again evaporated out of the chip, likely through the viewing window due to the microscope not being an ideal closed system. This second visit gave timelapse evidence that the HEK293 cells were surviving and beginning to multiply in the chip as highlighted below. The visit again sparked new approaches to trapping cells to be tested at a later date. Unfortunately, this visit occurred towards the end of iGEM, and further experiments will be conducted outside of iGEM. However, this visit provided definitive proof of mammalians cells surviving and dividing inside our low-cost, facilitated design microfluidic workflow.
{Attributions/Disclaimer: Present was Shane McCormack of the BU iGEM HW team, as well as Trinh Nguyen and mentor Brian Teague of the MIT iGEM team. All handling of biological materials performed by Trinh Nguyen of the MIT iGEM team and Brian Teague, mentor of the MIT iGEM team}
{Attributions/Disclaimer: Present was Shane McCormack of the BU iGEM HW team, as well as Trinh Nguyen and mentor Brian Teague of the MIT iGEM team. All handling of biological materials performed by Trinh Nguyen of the MIT iGEM team and Brian Teague, mentor of the MIT iGEM team}





Designing a Device to Characterize Starvation-Based Genetic Part
AUG - SEPT 2016 | A collaboration with the Northeastern 2016 iGEM Wetlab Team
We designed a microfluidic device for Northeastern University's iGEM team to help characterize their starvation-induced genetic part, by flowing cells through a set of cell traps placed after a gradient generator, then flowing two variable amounts of nutrients through the inputs of the gradient generator, causing a variable amount of nutrients to hit each cell trap, and cause a variable expression level of the starvation-linked fluorescence. Unfortunately, due to difficulties on both sides, the device and the cells to put through it were never realized. The preliminary sketches and designs are shown here, however.

Open Source Materials and Synthetic Biology
Open source materials encourage a cooperative and connected community in any field of research. It allows individuals and labs to easily share their ideas for others to build off of and can inspire even greater creations. So we teamed up with the BU 2016 iGEM Wetlab Team to investigate the current place of open source materials in Synthetic biology and how, moving forward, open source materials could impact the direction of the field. We recorded our findings biweekly through posts in a collaborative blog entitled "Who Owns What".

A General Overview of Intellectual Property in Synthetic Biology: PART 1
17 AUG | Blog Post 1
 Rachel Petherbridge, BostonU
Rachel Petherbridge, BostonU
An overview of the basics of intellectual property, which looks into background and definitions such as patent, copyright and trademark, as well as a roadmap to future posts.
 Read more
Read more

A General Overview of Intellectual Property in Synthetic Biology: PART 2
22 AUG | Blog Post 2
 Kestas Subacius, BostonU_HW
Kestas Subacius, BostonU_HW
Further discussion of general intellectual property practices while drawing comparisons between intellectual property in synthetic biology and the software.
 Read more
Read more


