| Line 677: | Line 677: | ||
$(window).load(function() { | $(window).load(function() { | ||
$(window).resize(); | $(window).resize(); | ||
| + | $('body').css('opacity', '1'); | ||
}) | }) | ||
| Line 789: | Line 790: | ||
| + | </script> | ||
| + | |||
| + | <script> | ||
| + | var clicked = ''; | ||
| + | var current = 'background'; // sets initial display to be purpose | ||
| + | $('.navigator').click( function() { // when any element with .class is clicked, trigger function | ||
| + | var selected = $(this).attr('data-select'); // grabs the name of the navigational element clicked | ||
| + | if (selected != current) { // only change display / trigger animation if clicking different nav | ||
| + | changeText(selected) | ||
| + | } | ||
| + | }) | ||
| + | |||
| + | var dir = location.hash; // grabs the anchor (text following the #) | ||
| + | var n = dir.indexOf('?'); // and initially sets the page | ||
| + | dir = dir.substring(0, n != -1 ? n : dir.length); | ||
| + | dir = dir.replace('#',''); | ||
| + | if (dir == '') { | ||
| + | dir = 'background'; | ||
| + | } | ||
| + | changeText(dir) | ||
| + | |||
| + | $(".subNav").click(function() { | ||
| + | clicked = $(this).attr('data-select'); | ||
| + | $('html, body').animate({ | ||
| + | scrollTop: $("#h4-"+ clicked).offset().top | ||
| + | }, 0000); | ||
| + | }); | ||
| + | |||
| + | |||
| + | function changeText(selected) { | ||
| + | $('.navigator').css('border-bottom','1px solid #A8A8A8'); // make all navigational element have no bottom border | ||
| + | $('#th-' + selected).css('border-bottom', '4px solid #A8A8A8'); // the clicked navigational element given border // hides all text | ||
| + | var widthText = $('.longText').outerWidth(false); | ||
| + | $('.textSection').css('width', widthText); | ||
| + | $('.textSection').css('display','none'); | ||
| + | $('.subNavList').css('display','none'); // slighly shifts right all text to set up for animation | ||
| + | $('.textSection').css('margin-left', '60px') // slighly shifts right all text to set up for animation | ||
| + | $('#div-' + selected).css('display','block'); // the div containing the text is displayed | ||
| + | $('#div-' + selected).animate({ marginLeft: '0px'}, 200); | ||
| + | $('.textSection').css('width', '100%'); | ||
| + | current = selected; // sets the current display | ||
| + | location.hash = current; | ||
| + | } | ||
| + | |||
</script> | </script> | ||
</html> | </html> | ||
Revision as of 07:06, 19 October 2016
</div> </div>

Background |
Pathway |
Optimization |
Results |
|---|
Background
The Modeling portion of the UMaryland iGEM project proposes specific adjustments to the expression levels of enzymes in the methane digestion pathways. Our modeling efforts focus on ensuring the viability of our engineered organism, and optimizing the efficiencies of our pathways. Along the way, our team encountered stumbling blocks that we would like to illuminate for iGEM teams in the future. Incorporated into our modeling page is a concise guide on getting started with the Matlab applet, Simbiology, to model simple metabolic pathways. If you have no interest in this guide, please feel free to optimize your time by skipping over the purple text.
Creating the Pathway Architecture
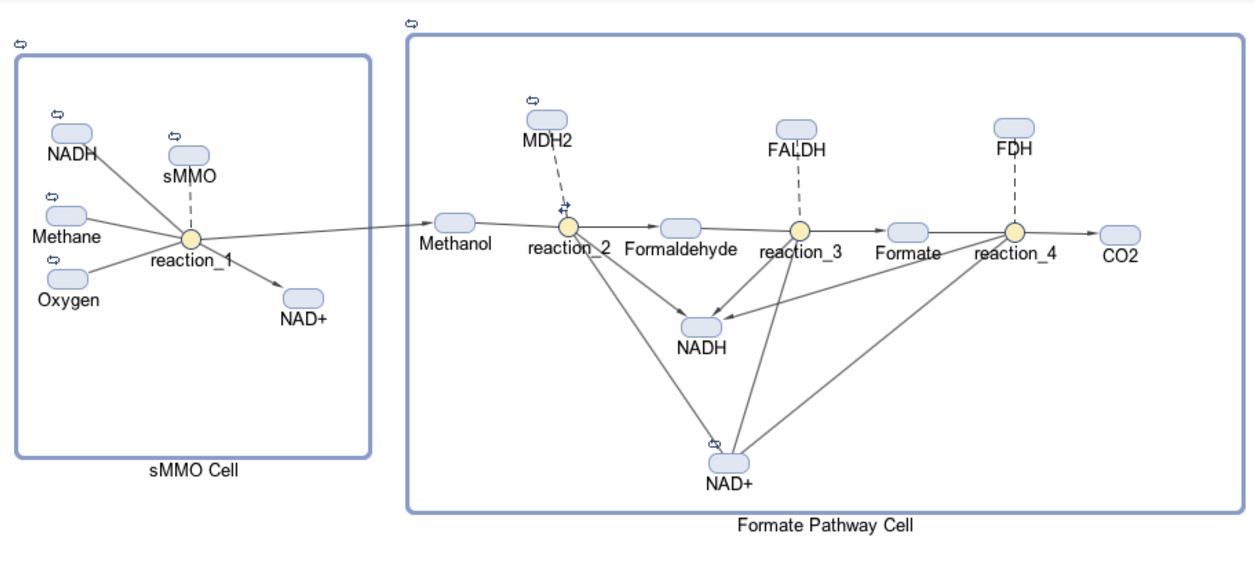
The models for our formate and fructose pathways each consist of two compartments, four reactions and 14 or 15 species, respectively. The initial goal for our project was to produce a co-culture of cells consisting of “sMMO Cells” that produce methanol and “Formate/Fructose Cells” that detoxify methanol, all in an effort to completely biodegrade methane. The first compartment in our model shown below is the “sMMO Cell” that converts methane to methanol, which travels to the second compartment of our model, either the “Formate” or “Fructose Cell.” Each species represents a different molecule involved in our pathway. The architecture for our Formate pathway displays a progression of compounds starting at methane and ending at carbon dioxide. In all, this pathway is facilitated by four enzymes.

The first half of each pathway is identical. The first enzyme used in each pathway is sMMO, which catalyses the oxidation of methane to methanol while simultaneously oxidizing NADH to NAD+. Both pathways then proceed to oxidize methanol to formaldehyde, which is a reaction catalyzed by the MDH2 enzyme.
Once formaldehyde is produced, the two pathways begin to differ. In the Formate Pathway, formaldehyde is first oxidized to formate, which is then oxidized to carbon dioxide by FALDH and FDH enzymes respectively. In the Fructose Pathway, formaldehyde is incorporated into a 5 carbon sugar by HPS to make D-arabinose 6-phosphate, a 6 carbon sugar. PHI then converts D-arabinose 6-phosphate into D-fructose-6-phosphate, which is a substrate of glycolysis.




