| Line 6: | Line 6: | ||
<head> | <head> | ||
<script> | <script> | ||
| + | $('body').css('display', 'none'); | ||
$('body').css('opacity', '0'); | $('body').css('opacity', '0'); | ||
</script> | </script> | ||
| Line 11: | Line 12: | ||
<script src="https://static.igem.org/mediawiki/2016/c/ce/T--UMaryland--parallax.txt"></script> | <script src="https://static.igem.org/mediawiki/2016/c/ce/T--UMaryland--parallax.txt"></script> | ||
<script src="https://static.igem.org/mediawiki/2016/d/d3/T--UMaryland--slick.txt" type="text/javascript"></script> | <script src="https://static.igem.org/mediawiki/2016/d/d3/T--UMaryland--slick.txt" type="text/javascript"></script> | ||
| − | <title> | + | <title>Human Practices</title> |
</head> | </head> | ||
| Line 290: | Line 291: | ||
} | } | ||
| − | #div-navResized.pushed a | + | #div-navResized.pushed a:hover { |
opacity: .7; | opacity: .7; | ||
text-decoration: none; | text-decoration: none; | ||
| Line 356: | Line 357: | ||
<style> | <style> | ||
| + | |||
.caption { | .caption { | ||
font-weight: bold; | font-weight: bold; | ||
| − | + | margin-left: 25px; | |
} | } | ||
| Line 368: | Line 370: | ||
right: auto; | right: auto; | ||
width: 80%; | width: 80%; | ||
| − | min-width: | + | min-width: 600px; |
max-width: 1500px; | max-width: 1500px; | ||
} | } | ||
| Line 408: | Line 410: | ||
padding-top: 50px; | padding-top: 50px; | ||
padding-bottom: 150px; | padding-bottom: 150px; | ||
| + | min-height: 800px; | ||
} | } | ||
| Line 443: | Line 446: | ||
h4 { | h4 { | ||
/* formats intratext section headings */ | /* formats intratext section headings */ | ||
| + | margin-top: 40px !important; | ||
font-size: 20pt; | font-size: 20pt; | ||
| − | |||
} | } | ||
| Line 459: | Line 462: | ||
.figure { | .figure { | ||
/* fixes the width of images in results tables */ | /* fixes the width of images in results tables */ | ||
| − | width: | + | width: 80%; |
display: block; | display: block; | ||
| + | margin-top: 20px; | ||
| + | margin-bottom: 20px; | ||
| + | margin-left: 7%; | ||
| + | margin-right: 13%; | ||
| + | } | ||
| + | |||
| + | .figureImg { | ||
| + | width: 100%; | ||
border-style: solid; | border-style: solid; | ||
border-color: #A8A8A8; | border-color: #A8A8A8; | ||
border-width: 2px; | border-width: 2px; | ||
| − | + | } | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
/* formats caption and heading of results tables */ | /* formats caption and heading of results tables */ | ||
| Line 502: | Line 509: | ||
#table-selector { | #table-selector { | ||
| − | width: | + | width: 80%; |
| − | + | min-width: 600px; | |
| − | + | max-width: 1500px; | |
| − | + | position: absolute; | |
border-collapse: collapse; | border-collapse: collapse; | ||
background: none; | background: none; | ||
| Line 511: | Line 518: | ||
#table-selector th { | #table-selector th { | ||
| + | width: 25%; | ||
text-align: center; | text-align: center; | ||
| − | |||
} | } | ||
| Line 529: | Line 536: | ||
th p { | th p { | ||
| − | font-size: | + | font-size: 16pt !important; |
font-weight: 400; | font-weight: 400; | ||
cursor: pointer; | cursor: pointer; | ||
| Line 536: | Line 543: | ||
} | } | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</style> | </style> | ||
| − | <nav> | + | <nav> |
| − | + | <a href="https://2016.igem.org/Team:UMaryland"><img src="https://static.igem.org/mediawiki/2016/2/26/T--UMaryland--newlogo.jpeg" id="img-logo" width="128px"></a> | |
| − | + | ||
| − | + | ||
<ul> | <ul> | ||
<a style="opacity: 0">Fix</a> | <a style="opacity: 0">Fix</a> | ||
| Line 560: | Line 559: | ||
<!-- Nav bar when more button is clicked --> | <!-- Nav bar when more button is clicked --> | ||
<div id="div-navResized"> | <div id="div-navResized"> | ||
| − | + | <img src="https://static.igem.org/mediawiki/2016/2/26/T--UMaryland--newlogo.jpeg" id="img-logoResized"> | |
<ul> | <ul> | ||
<li><a href="https://2016.igem.org/Team:UMaryland/projects">Projects</a></li> | <li><a href="https://2016.igem.org/Team:UMaryland/projects">Projects</a></li> | ||
| Line 570: | Line 569: | ||
</ul> | </ul> | ||
</div> | </div> | ||
| − | <div data-parallax="scroll" data-image-src="https://static.igem.org/mediawiki/2016/ | + | <div class="scrollContainer"> |
| − | + | <div data-parallax="scroll" data-image-src="https://static.igem.org/mediawiki/2016/a/a8/T--UMaryland--smmo.jpg" class="div-scrollPic" style="height: 650px;" > | |
| − | + | <div class="titleText"> | |
| − | <h11> | + | <style> |
| − | <h21> | + | </style> |
| − | <h31> | + | <!-- where background image below titles show --> |
| − | + | <h11>Modeling</h11></br> | |
| − | + | <h21>Predicting Data and Optimizing Results</h21></br> | |
| + | <h31>Applying engineering principals to biological systems</h31></br> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <!-- Heading text --> | ||
| + | |||
<div id="div-text"> <!-- Start of main text of page --> | <div id="div-text"> <!-- Start of main text of page --> | ||
<!-- Heading text --> | <!-- Heading text --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<style> | <style> | ||
| − | |||
| − | |||
| − | |||
| − | |||
.longText strong { | .longText strong { | ||
font-size: 14pt; | font-size: 14pt; | ||
| Line 603: | Line 596: | ||
} | } | ||
| + | .textSection { | ||
| + | display: none; | ||
| + | } | ||
| − | + | .longText p { | |
| − | + | margin-top: 15px; | |
| − | + | margin-bottom: 15px; | |
| − | + | } | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | .purple { | |
| − | + | color: #700A91; | |
| − | + | } | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</style> | </style> | ||
| − | < | + | <table id="table-selector"> |
| − | < | + | <thead> |
| − | + | <th class="borderRight navigator borderTop" id="th-background" data-select="background"><p>Background</p></th> | |
| − | < | + | <th class="borderBoth navigator borderTop" id="th-pathway" data-select="pathway"><p>Pathway</p></th> |
| − | < | + | <th class="borderBoth navigator borderTop" id="th-optimization" data-select="optimization"><p>Optimization</p></th> |
| − | + | <th class="borderLeft navigator borderTop" id="th-results" data-select="results"><p>Results</p></th> | |
| − | <p> | + | </thead> |
| + | </table> | ||
| + | </br></br></br> | ||
| + | <div class="longText"> | ||
| + | <div class="textSection" id="div-background"> | ||
| + | <h4>Background</h4> | ||
| + | <p>The Modeling portion of the UMaryland iGEM project proposes specific adjustments to the expression levels of enzymes in the methane digestion pathways. Our modeling efforts focus on ensuring the viability of our engineered organism, and optimizing the efficiencies of our pathways. Along the way, our team encountered stumbling blocks that we would like to illuminate for iGEM teams in the future. Incorporated into our modeling page is a concise guide on getting started with the Matlab applet, Simbiology, to model simple metabolic pathways. If you have no interest in this guide, please feel free to optimize your time by skipping over the purple text.</p> | ||
| + | <div class="purple"> | ||
| + | <p>Simbiology is a tool that enables teams with variable modeling backgrounds to build pathway architectures by using a simple drag and drop interface. Although Simbiology is an extremely useful and intuitive tool, it does require some experience to navigate, and it does have its errors; but more on that later. The first step is to open Simbiology by typing “simbiology” into the Matlab command window, then hitting enter.</p> | ||
| + | <p>To begin drawing your metabolic pathway, drag and drop a “reaction” icon, located on the left hand sign of the window, onto the blank canvas covering the right half of the window. Then double click on the small orange reaction circle that you placed, and enter your chemical reaction into the block property editor by replacing “null -> null” with your balanced formula. Enzymes are included in the balanced formula. Make sure your enzyme is represented as both a reactant and a product. Clicking out of the Block Property Editor will create species with arrows pointing into your reaction and arrows pointing out of your reaction. Enzymes are connected to the reaction by a dotted line, representing an intermediate that is not consumed or produced by the reaction. You may place as many reactions and species onto the canvas as you would like. If you would like an example model architecture, our pathways are shown later.</p> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
<div class="longText"> | <div class="longText"> | ||
| − | <h4> | + | <div class="textSection" id="div-pathway"> |
| − | + | <h4>Creating the Pathway Architecture</h4> | |
| + | <p>The models for our formate and fructose pathways each consist of two compartments, four reactions and 14 or 15 species, respectively. The initial goal for our project was to produce a co-culture of cells consisting of “sMMO Cells” that produce methanol and “Formate/Fructose Cells” that detoxify methanol, all in an effort to completely biodegrade methane. The first compartment in our model shown below is the “sMMO Cell” that converts methane to methanol, which travels to the second compartment of our model, either the “Formate” or “Fructose Cell.” Each species represents a different molecule involved in our pathway. The architecture for our Formate pathway displays a progression of compounds starting at methane and ending at carbon dioxide. In all, this pathway is facilitated by four enzymes.</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/7/75/T--UMaryland--formateModel.jpg" width="100%" /> | ||
| + | <p>The first half of each pathway is identical. The first enzyme used in each pathway is sMMO, which catalyses the oxidation of methane to methanol while simultaneously oxidizing NADH to NAD+. Both pathways then proceed to oxidize methanol to formaldehyde, which is a reaction catalyzed by the MDH2 enzyme.</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/e/eb/T--UMaryland--fructoseModel.jpg" width="100%" /> | ||
| + | <p>Once formaldehyde is produced, the two pathways begin to differ. In the Formate Pathway, formaldehyde is first oxidized to formate, which is then oxidized to carbon dioxide by FALDH and FDH enzymes respectively. In the Fructose Pathway, formaldehyde is incorporated into a 5 carbon sugar by HPS to make D-arabinose 6-phosphate, a 6 carbon sugar. PHI then converts D-arabinose 6-phosphate into D-fructose-6-phosphate, which is a substrate of glycolysis.</p> | ||
</div> | </div> | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </div | + | |
</div> | </div> | ||
<div id="bot-nav"> | <div id="bot-nav"> | ||
| Line 756: | Line 661: | ||
$('#div-slideshow').slick({ | $('#div-slideshow').slick({ | ||
| − | dots | + | dots: true |
| − | + | ||
| − | + | ||
}); | }); | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
$(window).resize(function() { | $(window).resize(function() { | ||
var marginTotal = $(window).outerWidth(true) - $('.titleText').outerWidth(false); | var marginTotal = $(window).outerWidth(true) - $('.titleText').outerWidth(false); | ||
| Line 794: | Line 676: | ||
var marginCheck = marginTitle / 2; | var marginCheck = marginTitle / 2; | ||
if (marginCheck < 15) { | if (marginCheck < 15) { | ||
| − | $('.titleText').css('left','15px'); | + | $('.titleText, #table-selector').css('left','15px'); |
| − | $('.titleText').css('margin-left','0px'); | + | $('.titleText, #table-selector').css('margin-left','0px'); |
} else { | } else { | ||
| − | $('.titleText').css('left','50%'); | + | $('.titleText, #table-selector').css('left','50%'); |
| − | $('.titleText').css('margin-left','-40%'); | + | $('.titleText, #table-selector').css('margin-left','-40%'); |
} | } | ||
}) | }) | ||
$(window).load(function() { | $(window).load(function() { | ||
| + | $(window).resize(); | ||
$('body').css('opacity', '1'); | $('body').css('opacity', '1'); | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}) | }) | ||
| Line 911: | Line 777: | ||
}); | }); | ||
</script> | </script> | ||
| − | <script> $('#iGEM').css('display','block'); </script> | + | <script> $('#iGEM').css('display','block'); |
| + | |||
| + | $('body').css('display', 'block');</script> | ||
<script> | <script> | ||
$(document).ready(function(){ | $(document).ready(function(){ | ||
| − | + | ||
| − | + | ||
| − | + | ||
}); | }); | ||
| Line 927: | Line 794: | ||
} | } | ||
}); | }); | ||
| − | |||
var toggle = 1; | var toggle = 1; | ||
| − | + | ||
| − | </script> | + | |
| + | </script> | ||
| + | |||
<script> | <script> | ||
var clicked = ''; | var clicked = ''; | ||
| − | var current = ' | + | var current = 'background'; // sets initial display to be purpose |
$('.navigator').click( function() { // when any element with .class is clicked, trigger function | $('.navigator').click( function() { // when any element with .class is clicked, trigger function | ||
var selected = $(this).attr('data-select'); // grabs the name of the navigational element clicked | var selected = $(this).attr('data-select'); // grabs the name of the navigational element clicked | ||
| Line 948: | Line 816: | ||
dir = dir.replace('#',''); | dir = dir.replace('#',''); | ||
if (dir == '') { | if (dir == '') { | ||
| − | dir = ' | + | dir = 'background'; |
} | } | ||
changeText(dir) | changeText(dir) | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
$(".subNav").click(function() { | $(".subNav").click(function() { | ||
| Line 973: | Line 827: | ||
}); | }); | ||
| − | |||
| − | |||
| − | |||
function changeText(selected) { | function changeText(selected) { | ||
$('.navigator').css('border-bottom','1px solid #A8A8A8'); // make all navigational element have no bottom border | $('.navigator').css('border-bottom','1px solid #A8A8A8'); // make all navigational element have no bottom border | ||
| − | $('#th-' + selected).css('border-bottom', '4px solid #A8A8A8'); // the clicked navigational element given border | + | $('#th-' + selected).css('border-bottom', '4px solid #A8A8A8'); // the clicked navigational element given border // hides all text |
| − | $('. | + | var widthText = $('.longText').outerWidth(false); |
| − | $('.subNavList').css('display','none'); // | + | $('.textSection').css('width', widthText); |
| − | $('. | + | $('.textSection').css('display','none'); |
| − | $('#div-' + selected).css('display','block'); | + | $('.subNavList').css('display','none'); // slighly shifts right all text to set up for animation |
| − | + | $('.textSection').css('margin-left', '60px') // slighly shifts right all text to set up for animation | |
| − | $('#div-' + selected).animate({ marginLeft: '0px'}, 200); | + | $('#div-' + selected).css('display','block'); // the div containing the text is displayed |
| + | $('#div-' + selected).animate({ marginLeft: '0px'}, 200); | ||
| + | $('.textSection').css('width', '100%'); | ||
current = selected; // sets the current display | current = selected; // sets the current display | ||
location.hash = current; | location.hash = current; | ||
Revision as of 07:50, 19 October 2016
</div> </div>

Background |
Pathway |
Optimization |
Results |
|---|
Background
The Modeling portion of the UMaryland iGEM project proposes specific adjustments to the expression levels of enzymes in the methane digestion pathways. Our modeling efforts focus on ensuring the viability of our engineered organism, and optimizing the efficiencies of our pathways. Along the way, our team encountered stumbling blocks that we would like to illuminate for iGEM teams in the future. Incorporated into our modeling page is a concise guide on getting started with the Matlab applet, Simbiology, to model simple metabolic pathways. If you have no interest in this guide, please feel free to optimize your time by skipping over the purple text.
Simbiology is a tool that enables teams with variable modeling backgrounds to build pathway architectures by using a simple drag and drop interface. Although Simbiology is an extremely useful and intuitive tool, it does require some experience to navigate, and it does have its errors; but more on that later. The first step is to open Simbiology by typing “simbiology” into the Matlab command window, then hitting enter.
To begin drawing your metabolic pathway, drag and drop a “reaction” icon, located on the left hand sign of the window, onto the blank canvas covering the right half of the window. Then double click on the small orange reaction circle that you placed, and enter your chemical reaction into the block property editor by replacing “null -> null” with your balanced formula. Enzymes are included in the balanced formula. Make sure your enzyme is represented as both a reactant and a product. Clicking out of the Block Property Editor will create species with arrows pointing into your reaction and arrows pointing out of your reaction. Enzymes are connected to the reaction by a dotted line, representing an intermediate that is not consumed or produced by the reaction. You may place as many reactions and species onto the canvas as you would like. If you would like an example model architecture, our pathways are shown later.
Creating the Pathway Architecture
The models for our formate and fructose pathways each consist of two compartments, four reactions and 14 or 15 species, respectively. The initial goal for our project was to produce a co-culture of cells consisting of “sMMO Cells” that produce methanol and “Formate/Fructose Cells” that detoxify methanol, all in an effort to completely biodegrade methane. The first compartment in our model shown below is the “sMMO Cell” that converts methane to methanol, which travels to the second compartment of our model, either the “Formate” or “Fructose Cell.” Each species represents a different molecule involved in our pathway. The architecture for our Formate pathway displays a progression of compounds starting at methane and ending at carbon dioxide. In all, this pathway is facilitated by four enzymes.
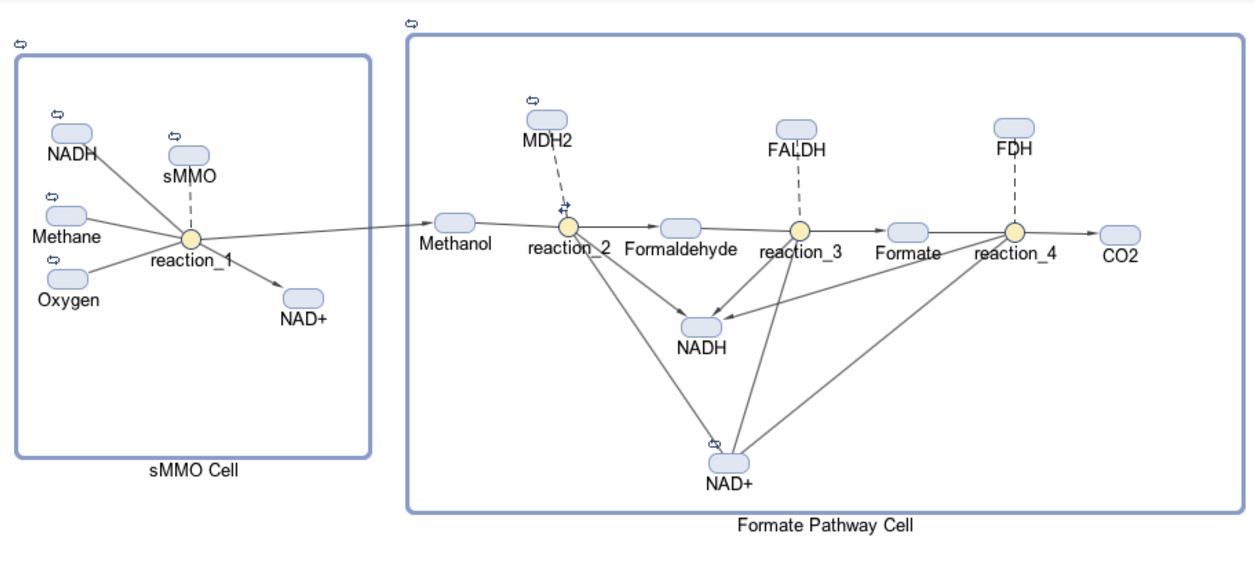
The first half of each pathway is identical. The first enzyme used in each pathway is sMMO, which catalyses the oxidation of methane to methanol while simultaneously oxidizing NADH to NAD+. Both pathways then proceed to oxidize methanol to formaldehyde, which is a reaction catalyzed by the MDH2 enzyme.

Once formaldehyde is produced, the two pathways begin to differ. In the Formate Pathway, formaldehyde is first oxidized to formate, which is then oxidized to carbon dioxide by FALDH and FDH enzymes respectively. In the Fructose Pathway, formaldehyde is incorporated into a 5 carbon sugar by HPS to make D-arabinose 6-phosphate, a 6 carbon sugar. PHI then converts D-arabinose 6-phosphate into D-fructose-6-phosphate, which is a substrate of glycolysis.




