| Line 537: | Line 537: | ||
</html> | </html> | ||
| − | Last year, we devised Flavorator which preserve food by fragrance component with antibacterial effect and introduced | + | Last year, we devised Flavorator which preserve food by fragrance component with antibacterial effect and introduced its concept. We chose geraniol and farnesol as the fragrance having an antibacterial effect, and we made E.coli possible produce these fragrance components by recombination. However, the synthetic amount of geraniol and farnesol was lower than expected, and it is not sufficient for practical use. |
| − | + | This year, to improve functionality and feasibility of Flavorator, we focused on the production of farnesol instead of two fragrances for three reasons. | |
| + | 1. Farnesol does not impair flavor of food because it is almost odorless. | ||
| + | 2. Farnesol has a high antifungal activity than others. | ||
| + | 3. E.coli already possesses synthetic pathway for farnesol so that it is easier to modify for over-expression of farnesol using authentic genes. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[File:経路図2.png|800px|thumb|left|Fig.1 Pathway of our design | [[File:経路図2.png|800px|thumb|left|Fig.1 Pathway of our design | ||
DXP: 1-Deoxy-D-xylulose 5-phosphate. MEP: 2-C-methylerythritol 4-phosphate. CDP-ME: 4-diphosphocytidyl-2-C-methylerythritol. CDP-MEP: 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate. MEC: 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate. HMBPP: (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate. IPP: Isopentenyl pyrophosphate. DMAPP: Dimethylallyl pyrophosphate. GPP: geranyl diphosphate. FPP: farnesyl diphosphate.]] | DXP: 1-Deoxy-D-xylulose 5-phosphate. MEP: 2-C-methylerythritol 4-phosphate. CDP-ME: 4-diphosphocytidyl-2-C-methylerythritol. CDP-MEP: 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate. MEC: 2-C-methyl-D-erythritol 2, 4-cyclodiphosphate. HMBPP: (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate. IPP: Isopentenyl pyrophosphate. DMAPP: Dimethylallyl pyrophosphate. GPP: geranyl diphosphate. FPP: farnesyl diphosphate.]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | To increase farnesol production, we improved farnesol synthetic process by adding three genes to E.coli from the last year. | |
| + | 1, The gene that increases the intermediate product of farnesol synthesis. | ||
| + | As 1-deoxy-D-xylulose 5-phosphate (DXP), a key gene for farnesol synthesis, is also used for another compound synthesis, we added a new DXP synthase to increase the intermediate product. | ||
| + | 2, The dephosphorylation enzyme gene | ||
| + | Last year, a synthesis of farnesol relied on endogenous dephosphorylation enzyme of E. coli, and this could be a bottleneck for farnesol production. This year, we introduced an additional endogenous phosphatase gene (YbjG, PgpB) in E. coli to increase the production of farnesol. | ||
| + | 3, A gene to improve resistance against farnesol. | ||
| + | As farnesol is an antibacterial fragrance, it is toxic to E.coli itself. So, we introduced the endogenous activator gene of AcrAB-TolC efflux pump, marA to improve resistance against farnesol. | ||
| − | E. coli has a pathway | + | Also, to improve the feasibility of Flavorator in the real situation, we thought that it is important to produce farnesol by non-recombinant E.coli. Therefore we attempted to increase production of farnesol using CRISPER / Cas9 system. |
| − | We tried to knockout gdhA | + | E. coli has their pathway to produce the farnesol. However, the gdhA gene, which is not lethal to E.coli, produce a byproduct from farnesol synthetic pathway and decrease the production of farnesol. We tried to knockout gdhA with CRISPER-CAS9. |
[[File:経路図3.png|800px|thumb|left|Fig.2 Pathway of our design | [[File:経路図3.png|800px|thumb|left|Fig.2 Pathway of our design | ||
Revision as of 03:20, 20 October 2016
Last year, we devised Flavorator which preserve food by fragrance component with antibacterial effect and introduced its concept. We chose geraniol and farnesol as the fragrance having an antibacterial effect, and we made E.coli possible produce these fragrance components by recombination. However, the synthetic amount of geraniol and farnesol was lower than expected, and it is not sufficient for practical use.
This year, to improve functionality and feasibility of Flavorator, we focused on the production of farnesol instead of two fragrances for three reasons. 1. Farnesol does not impair flavor of food because it is almost odorless. 2. Farnesol has a high antifungal activity than others. 3. E.coli already possesses synthetic pathway for farnesol so that it is easier to modify for over-expression of farnesol using authentic genes.
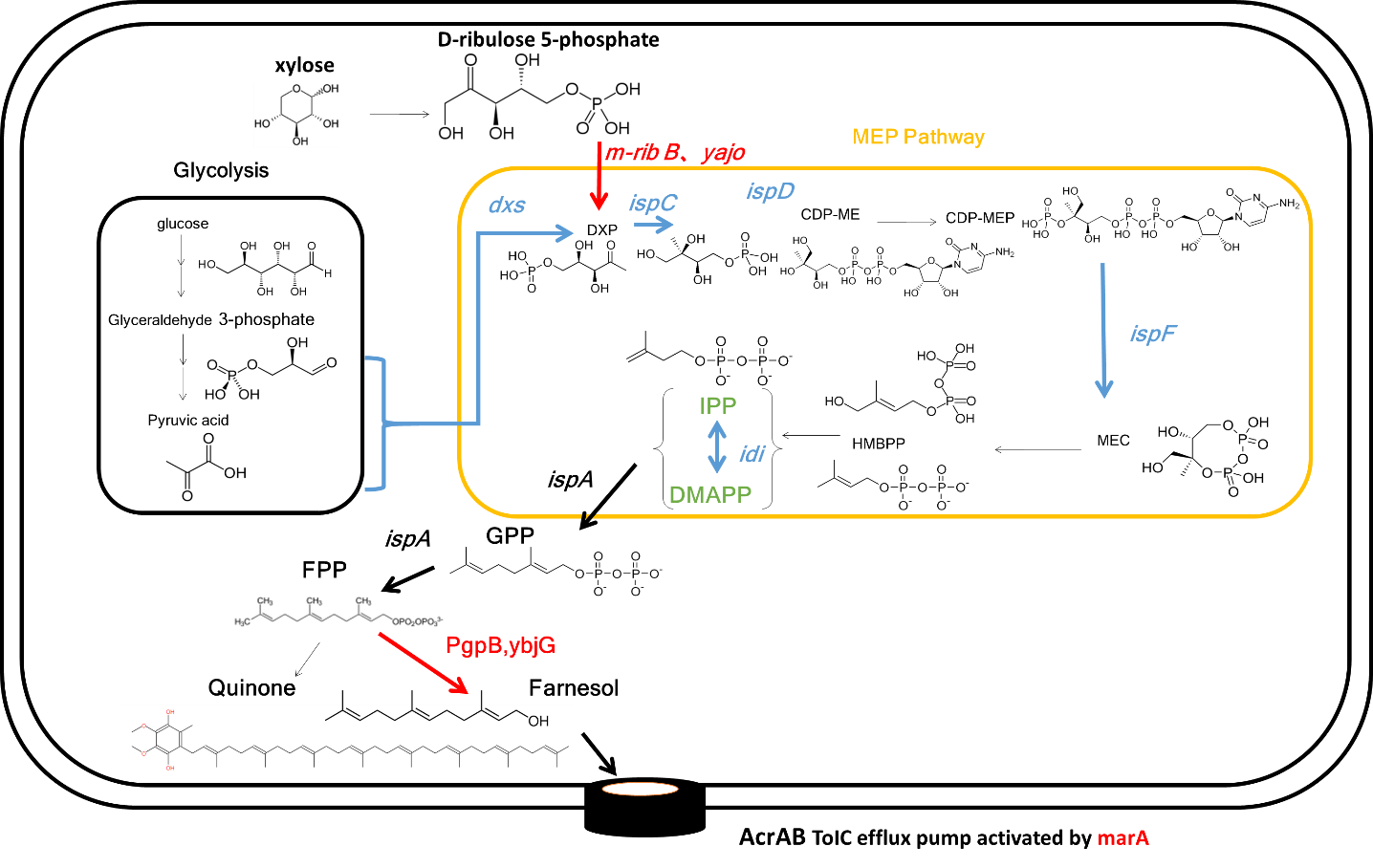
To increase farnesol production, we improved farnesol synthetic process by adding three genes to E.coli from the last year. 1, The gene that increases the intermediate product of farnesol synthesis. As 1-deoxy-D-xylulose 5-phosphate (DXP), a key gene for farnesol synthesis, is also used for another compound synthesis, we added a new DXP synthase to increase the intermediate product. 2, The dephosphorylation enzyme gene Last year, a synthesis of farnesol relied on endogenous dephosphorylation enzyme of E. coli, and this could be a bottleneck for farnesol production. This year, we introduced an additional endogenous phosphatase gene (YbjG, PgpB) in E. coli to increase the production of farnesol. 3, A gene to improve resistance against farnesol. As farnesol is an antibacterial fragrance, it is toxic to E.coli itself. So, we introduced the endogenous activator gene of AcrAB-TolC efflux pump, marA to improve resistance against farnesol.
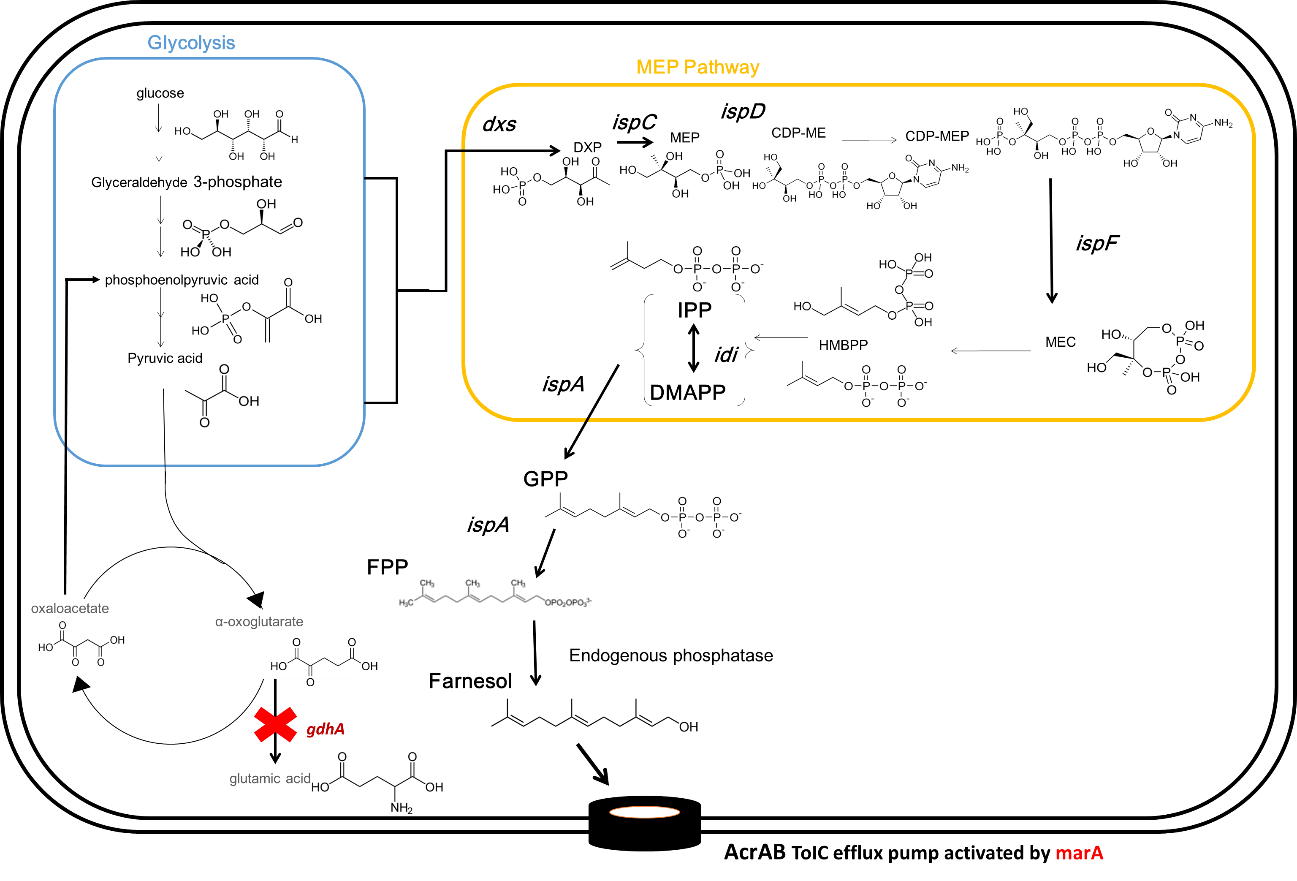
Also, to improve the feasibility of Flavorator in the real situation, we thought that it is important to produce farnesol by non-recombinant E.coli. Therefore we attempted to increase production of farnesol using CRISPER / Cas9 system. E. coli has their pathway to produce the farnesol. However, the gdhA gene, which is not lethal to E.coli, produce a byproduct from farnesol synthetic pathway and decrease the production of farnesol. We tried to knockout gdhA with CRISPER-CAS9.
REFERENCES
[http://onlinelibrary.wiley.com/doi/10.1002/biot.201600250/abstract?systemMessage=Wiley+Online+Library+will+be+unavailable+on+Saturday+30th+July+2016+from+08:00-11:00+BST+/+03:00-06:00+EST+/+15:00-18:00+SGT+for+essential+maintenance.Apologies+for+the+inconvenience [1] Wang C, Park JE, Choi ES, Kim SW.(2016).Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway – application of PgpB and YbjG phosphatases. Biotechnology Journal 11(10): 1291–1297.
[http://aem.asm.org/content/81/1/130.short [2] Kirby J, Nishimoto M, Chow W. N.R, Baidoo E.E., Wang G, Martin J, … & Keasling J D.(2015).Enhancing Terpene Yield from Sugars via Novel Routes to 1-Deoxy-d-Xylulose 5-Phosphate.Applied and Environmental Microbiology,81(1), 130-138.
[http://link.springer.com/article/10.1007/s00284-009-9408-9 [3]Gomes FIA, Teixeira P, Azeredo J, Oliveira R. (2009). Effect of Farnesol on Planktonic and Biofilm Cells of Staphylococcus epidermidis. Current Microbiology, 59(2), 118-122.
[http://www.nature.com/nbt/journal/v31/n3/full/nbt.2508.html%3FWT.ec_id%3DNBT-201303 [4]Jian W, Bikard D, Cox D, Zhang F & Marraffini LA. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31(3), 233-239.
[http://www.sciencedirect.com/science/article/pii/S1096717615000750 [5]Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang Y, Chen T, Zhao X.(2015). Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metabolic Engineering, 31, 13-21.
[http://www.sciencedirect.com/science/article/pii/S1389172312004185 [6]Shah A A, Wang C, Chung Y R, Kim J Y, Choi E S, Kim S W. (2013).Enhancement of geraniol resistance of Escherichia coli by MarA overexpression. Journal of Bioscience and Bioengineering, 115(3),235–258.
[http://nar.oxfordjournals.org/content/early/2016/04/08/nar.gkw223.abstract [7]Cui L, Bikard D. (2016).Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic acids research, gkw223.
[http://www.sciencedirect.com/science/article/pii/S1096717605000741 [8]Yuan L.Z, Rouvière P.E, LaRossa R.A, Suh W. (2006).Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metabolic Engineering 8(1) 79–90.
[http://aem.asm.org/content/81/15/5103.short [9]Pyne ME, Moo-Young M, Chung DA, Chou CP. 2015. Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli. Applied and Environmental Microbiology 81:5103–5114.

