| Line 435: | Line 435: | ||
<script> | <script> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<script> | <script> | ||
$('#page-contents a').click(function() { | $('#page-contents a').click(function() { | ||
Revision as of 06:31, 8 November 2016
Contents
- 1 Protocol of KOD PCR
- 1.1 LB media: (performed 3 times to make 3L total)
- 1.2 LB agar: (performed 3 times to make 3L total)
- 1.3 Bt Protein Extraction Procedures
- 1.4 SDS-PAGE
- 1.5 Agarose gel electrophoresis
- 1.6 Bacterial DNA Extraction
- 1.7 In-fusion
- 1.8 Plasmid Extraction
- 1.9 Protocol of C. reinhardtii Transformation
- 1.10 TAP Medium and C. reinhardtii Culture
- 1.11 Protocol of Mosquitoes (Larvae) Feeding
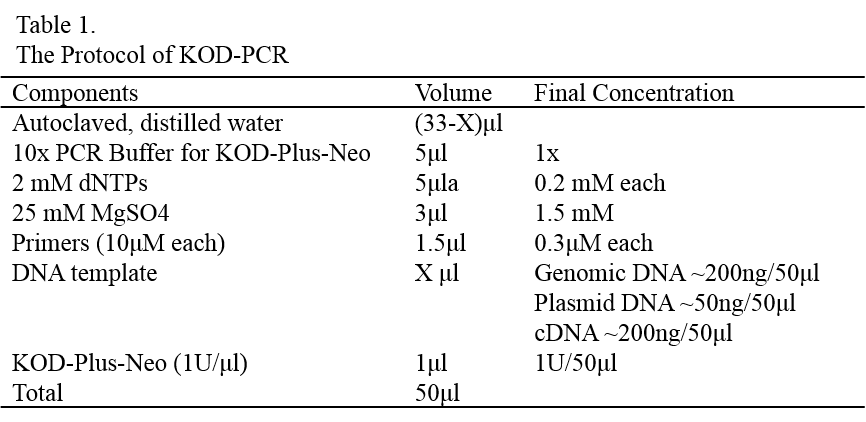
Protocol of KOD PCR
LB media: (performed 3 times to make 3L total)
Ingredients: Tryptone 10g, Yeast extract 5g, NaCl 10g, MilliQ water to 1000ml.
Methods: Dissolved 10g tryptone, 5g yeast extract and 10g NaCl in 800 mL MilliQ water, making use of a magnetic stirrer. Once dissolved, brought volume up to 1L using MilliQ water. Autoclaved 1000ml of the solution (121°C, 15 min, standard liquid cycle).
LB agar: (performed 3 times to make 3L total)
Ingredients: LB media broth 1000ml, Bacto agar 15g.
Methods: Added the 15 Bacto agar to 1000 mL of LB media and autoclave [121°C, 15 min, standard liquid cycle]. Add 1000µl of Chloroamphenicol (25 mg/mL), Ampicillin (50 mg/mL) or Kanamycin (30 mg/mL) and mixed well before plating out and setting agar. After cooling and antibiotic addition, the LB agar was plated out using aseptic technique. In total, 31 Ampicillin LB agar plates, 33 Chloramphenicol LB agar plates & 32 Kanamycin LB Agar plates resulted. All plates were aseptically sealed using parafilm and stored in a refrigerator. Ultimately, this procedure resulted in three 1L LB agar solutions with a different antibiotic in each.
Bt Protein Extraction Procedures
Prior to protein extraction pre-chill the protein extraction filter cartridge with collection tube on ice.
Pellet cells from bacterial culture in a 1.5-2.0 ml microcentrifuge tube with a table-top microcentrifuge at top speed for 1-2 min. Remove the supernatant and wash the cell pellets in cold PBS once. Aspirate the supernatant and leave small amount of PBS (about equivalent volume of packed cells) in the tube. Vortex briefly to resuspend the cells.
Add appropriate amounts of room temperature bacterial lysis buffer A to cell suspension (Table 1). Vortex vigorously for 10 second.
Note: presence of small amount of unlysed cells would not affect the quality of the samples.
Add appropriate amounts of buffer B (1/10 of buffer A) to the lysate and vortex vigorously for 10 seconds.
Transfer/pour the cell lysate to pre-chilled filter cartridge, and centrifuge in a microcentrifuge at 14,000-16,000 rpm for 30 seconds.
The clear cell lysate in collection tube is ready to use. Discard the filter cartridge according to your institution's waste disposal protocol. The protein yield with this method is about 2.0-3.0 mg/ml.
SDS-PAGE
Separation Gel
4.5 mL Acrylamid (40 %)
3.75 mL Tris-buffer pH 8.8
6.45 mL H20
0.15 mL SDS (10 %)
6 µL TEMED
0.15 mL APS (10 %)
Stacking gel
0.75 mL Acrylamid (40 %)
0.76 mL Tris-buffer pH 8.8
4.37 mL H20
0.2 mL SDS (10 %)
6 µL TEMED
60 µL APS (10 %)
Procedure
Mix all the reagents, adding APS and TEMED last (gives 2 gels).
Fill the separation gel in the SDS page apparatus and overlay with isopropanol.After polymerization discard the isopropanol and fill the stacking gel on top and insert the comb.
Preparation of the samples (add Laemmli-buffer).
Load the gel and run at 200 V.
Agarose gel electrophoresis
Reagents
1.0 g Agarose
100 mL 1x TAE buffer
4 µL Midori Green solution
x µL 6x Loading dye
5 µL DNA-Ladder Mix
5-15 µL DNA sample
Procedure
Add agarose to TAE buffer and heat in microwave until the agarose is completely dissolved. Cool down the solution until it is about 40 °C and pour it in the gel electrophoresis apparatus. Add 4 μl Midori Green to the poured gel, mix the suspension and add the comb. Place the solid gel in the gel electrophoresis apparatus (with the wells near the cathode) and fill with 1x TAE buffer. Fill the wells with desired amount of DNA mixed with the 6x loading dye (to end up with a 1x loading dye).
Start the gel electrophoresis by applying 100-120 V and stop when marker reaches the bottom of the gel.
Bacterial DNA Extraction
1. Culture bacteria in LB media to log-phase. (Overnight culture can be used in many cases.)
2. Centrifuge no more than 3 mL culture or 1 x 109 cells at 4,000 x g for 10 minutes at room temperature.
3. Aspirate and discard the media.
4. Add 100 μL TE Buffer. Vortex to completely resuspend the pellet.
5. Add 10 μL Lysozyme.
6. Incubate at 37°C for 10 minutes.
Note: The amount of enzyme required and/or the length of incubation may need to be modified depending on the bacterial strain used. Complete digestion of the cell wall is essential for efficient lysis. Longer incubation time may yield better results.
Optional: Follow the short protocol below for difficult to lyse bacteria.
1. Add 25 mg glass beads to 1.5 mL microcentrifuge tube.
2. Add sample to the glass beads.
3. Vortex at maximum speed for 5 minutes.
4. Let sample stand to allow the beads to settle.
5. Transfer supernatant to a new 1.5 mL microcentrifuge tube.
7. Add 100 μL BTL Buffer and 20 μL Proteinase K Solution. Vortex to mix thoroughly.
8. Incubate at 55°C in a shaking water bath.
Note: Usually no more than 1 hour is required for bacterial lysis. If a shaking water bath is not available, incubate the samples and shake or briefly vortex every 20-30 minutes.
9. Add 5 μL RNase A. Invert tube several times to mix.
10. Incubate at room temperature for 5 minutes.
11. Centrifuge at 10,000 x g for 2 minutes to pellet any undigested material.
12. Transfer the supernatant to a new 1.5 mL microcentrifuge tube. Do not disturb the pellet.
13. Add 220 μL BDL Buffer. Vortex to mix.
14. Incubate at 65°C for 10 minutes.
Note: A wispy precipitate may form upon addition of BDL Buffer; it does not interfere with DNA recovery.
15. Add 220 μL 100% ethanol. Vortex for 20 seconds at maximum speed to mix thoroughly.
Note: If any precipitate can be seen at this point, break the precipitate by pipetting up and down 10 times.
16. Insert a HiBind® DNA Mini Column into a 2 mL Collection Tube.
17. Transfer the entire sample to the HiBind® DNA Mini Column, including any precipitate that may have formed.
18. Centrifuge at 10,000 x g for 1 minute.
19. Discard the filtrate and the collection tube.
20. Insert the HiBind® DNA Mini Column into a new 2 mL Collection Tube.
21. Add 500 μL HBC Buffer.
Note: HBC Buffer must be diluted with isopropanol before use. Please see Page 4 for instructions.
22. Centrifuge at 10,000 x g for 1 minute.
23. Discard the filtrate and reuse the collection tube.
24. Add 700 μL DNA Wash Buffer.
Note: DNA Wash Buffer must be diluted with 100% ethanol before use. Please see Page 4 for instructions.
25. Centrifuge at 10,000 x g for 1 minute.
26. Discard the filtrate and reuse the collection tube.
27. Repeat Steps 24-26 for a second DNA Wash Buffer wash step.
28. Centrifuge the empty HiBind® DNA Mini Column at maximum speed (≥10,000 x g) for 2 minutes to dry the column.
Note: This step is critical for removal of trace ethanol that may interfere with downstream applications.
29. Insert the HiBind® DNA Mini Column into a new nuclease-free 1.5 mL microcentrifuge tube.
30. Add 50-100 μL Elution Buffer heated 65°C to the HiBind® DNA Mini Column.
Note: Make sure to add the Elution Buffer to the center of the HiBind® matrix. Each 50-100 μL elution typically yields 60-70% of the DNA bound to the HiBind® matrix. Two elutions generally yield ~90%. However, increasing elution volume reduces the concentration of the final product. To obtain DNA at higher concentrations, elution can be carried out using 50 μL Elution Buffer (which slightly reduces overall DNA yield). Volumes lower than 50 μL greatly reduce yields.
31. Let sit for 3 to 5 minutes at room temperature.
Note: Yields may be increased by incubating the column at 65°C (rather than at room temperature).
32. Centrifuge at 10,000 x g for 1 minute to elute the DNA.
33. Repeat Steps 30-32 for a second elution step.
34. Store eluted DNA at -20°C.
In-fusion
1. Set up the In-Fusion cloning reaction
10ul | Total Volume |
2 μl | 5X In-Fusion HD Enzyme Premix |
x μl* | Linearized Vector |
x μl* | Purified PCR Fragment |
x μl | dH2O (as needed) |
* For reactions with larger volumes of vector and PCR insert (> 7 μl of vector + insert), double the amount of enzyme premix, and add dH20 for a total volume of 20 μl.
2. Incubate the reaction for 15 min at 50 °C, then place on ice.
3. Continue to the Transformation Procedure. You can store the cloning reactions at –20°C until you are ready.
Plasmid Extraction
We use E.Z.N.A. Plasmid DNA Mini Kit I bought from OMEGA bio-tek company to
extract plasmids and the detail protocol is showed as bellow:
1. Isolate a single colony from a freshly streaked selective plate, and inoculate a culture of 1- 5 mL LB medium containing the appropriate selective antibiotic. Incubate for ~12-16 hours at
37°C with vigorous shaking (~ 300 rpm).
2. Centrifuge at 10,000 x g for 1 minute at room temperature
3. Decant or aspirate and discard the culture media.
4. Add 250 mL Solution I/RNase A. Vortex or pipet up and down to mix thoroughly.
5. Transfer suspension into a new 1.5 mL microcentrifuge tube.
6. Add 250 mL Solution II. Invert and gently rotate the tube several times to obtain a clear lysate. A 2-3 minute incubation may be necessary.(Avoid vigorous mixing)
7. Add 350 mL Solution III. Immediately invert several times until a flocculent white precipitate forms.
8. Centrifuge at maximum speed (≥13,000 x g) for 10 minutes. A compact white pellet will form. Promptly proceed to the next step.
9. Insert a HiBind DNA Mini Column into a 2 mL Collection Tube.
10. Transfer the cleared supernatant from Step 8 by CAREFULLY aspirating it into the HiBind DNA Mini Column. Be careful not to disturb the pellet and that no cellular debris is transferred to the HiBind DNA Mini Column.
11. Centrifuge at maximum speed for 1 minute.
12. Discard the filtrate and reuse the collection tube.
13. Add 500 mL HB Buffer.
14. Centrifuge at maximum speed for 1 minute.
15. Discard the filtrate and reuse collection tube.
16. Add 700 mL DNA Wash Buffer.
17. Centrifuge at maximum speed for 1 minute.
18. Discard the filtrate and reuse the collection tube.
19. Centrifuge the empty HiBind DNA Mini Column for 2 minutes at maximum speed to dry the column matrix.
20. Transfer the HiBind DNA Mini Column to a clean 1.5 mL microcentrifuge tube.
21. Add 30-100 μl Elution Buffer or sterile deionized water directly to the center of the column membrane.
22. Let sit at room temperature for 1 minute.
23. Centrifuge at maximum speed for 1 minute.
24. Store DNA at -20°C.
Protocol of C. reinhardtii Transformation
1. Measure the optical density of the C. reinhardtii cultures at 750nm (OD750)
Note: For best performance, the OD750 of cultures should be between 0.3–0.5. If the OD750 does not reach 0.3 within 24 hours of incubation after dilution, incubate the cells for an extra 3–5 hours to allow for an additional cell division.
2. Harvest 15mL of the cell (per transformation) by centrifugation at 2,500 rpm for 10 minutes at room temperature. Centrifuge the cells in 15-Ml conical tubes to obtain tight pellets.
3. Discard the supernatant by decanting. Remove the remaining supernatant using a pipette.
4. Resuspend the cells in 250 μL of TAP-40 mM sucrose solution at room temperature by gently pipetting up and down.
5. Add 2 μg linearized plasmid DNA (i.e., pChlamy_3 construct containing your gene of interest) into the resuspended cells. Mix the DNA-cell suspension gently by flicking the tube. In a separate tube, prepare a control transformation with the pChlamy_2/Control Vector, linearized using PvfI restriction enzyme.
6. Transfer 250 μL of the transformation mixture into an electroporation cuvette and incubate at room temperature for 5 minutes.
7. While the transformation mixtures are incubating, add 5 mL of TAP-40 mM sucrose solution at room temperature into each well of a 6-well plate.
Note: You will divide each transformation mixture between two wells of a 6-well plate after electroporation, so that the cells in each transformation mixture will recover in 10 mL of TAP-40 mM sucrose solution total.
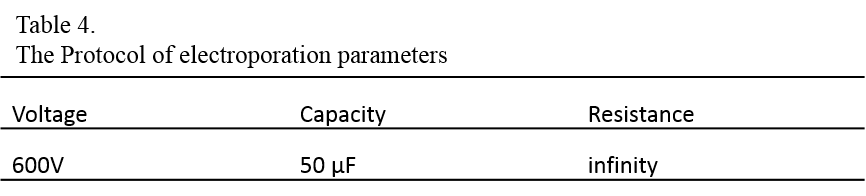
8. Set the electroporation parameters as follows:
9. Gently tap the electroporation cuvette to mix the contents and resuspend the settled cells, and place the cuvette in the cuvette chamber.
10. Electroporate the cells with the above parameters.
11. Split the transformation mixture into two aliquots of 125 μL each and transfer each aliquot into one well of the 6-well plate containing 5 mL/well of TAP-40 mM sucrose solution at room temperature. Wash the cuvette with 1 mL of TAP-40 mM sucrose solution to get most cells out of the cuvette and split and add the wash into the same two wells.
12. Place the 6-well plate in the algal growth chamber set to 26°C and 50 μE m–2 s–1.
13. Incubate the cells for 24 hours with gentle agitation (100–150 rpm) to let them recover.
14. Centrifuge the cells at 2,500 rpm for 10 minutes at room temperature.
15. Discard the supernatant by decanting. Remove the remaining supernatant with a pipette.
16. Resuspend the cells with gentle pipetting in 150 μL of TAP-40 mM sucrose solution at room temperature.
17. Plate the entire cell solution from each transformation on one TAP-agar-Hygromycin plate using disposable cell spreaders or glass plating beads to spread the cells evenly. Make sure the plates do not have condensation on them.
18. Place the plates agar side at the bottom in algal growth chamber set to 26°C and 50 μE m–2 s–1. Do not stack the plates to ensure continuous and even illumination.
19. Incubate the plates for 5 days or until C. reinhardtii colonies are clearly visible. Control vector should produce a minimum of 30 transform ants per electroporation reaction. The transformation efficiency with the pChlamy_3 construct will depend on the nature, size, and codon content of the gene of interest, and the physiological state of the cells.
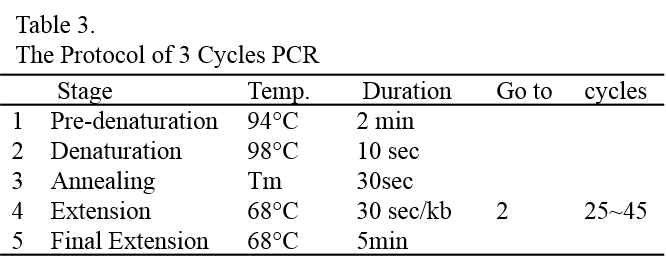
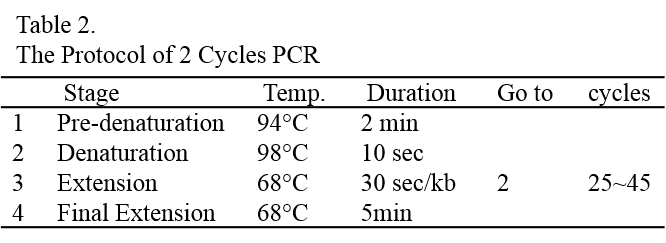
20. Proceed to determination of integration by colony PCR (see page 16) before selecting clones for further scale-up. About 20% of the colonies should be positive for the gene of interest. Due to random integration and silencing events in C. reinhardtii, we recommend picking at least 10 positive colonies and testing them for the expression level of the gene of interest by RT-PCR (or Western blotting, if you have the antibody to detect it).
TAP Medium and C. reinhardtii Culture
Tris Acetate Phosphate (TAP) medium from Gorman, D.S., and R.P. Levine (1965) Proc. Natl. Acad. Sci. USA 54, 1665-1669.
This is probably the most widely-used medium at present for experimental work.
Following solutions are prepared.
1. TAP salts
NH4Cl 15.0 g
MgSO4.7H2O 4.0 g CaCl2.2H2O 2.0 g
water to 1 liter
2. phosphate solution
K2HPO4 28.8 g
KH2PO4 14.4 g
water to 100 ml
3. Hutner's trace elements
To make the final medium, mix the following:
2.42 g Tris
25 ml solution #1 (salts)
0.375 ml solution #2 (phosphate)
1.0 ml solution #3 (trace elements)
1.0 ml glacial acetic acid
water to 1 liter
For solid medium, add 15 g agar per liter
Autoclave.
For Tris-minimal medium omit the acetic acid and titrate the final solution to pH 7.0 with HCl
Reference:http://www.chlamy.org/TAP.html
Hutner's trace elements
Hutner et al. (1950) Proc. Am. Philos. Soc. 94, 152-170
This mixture is used both in TAP and in the Sueoka high salt medium.
For a detailed analysis of how well this trace elements solution meets the nutritional requirements of C. reinhardtii,
see Merchant et al. (2006) Biochim. Biophys. Acta 1763, 578-594.
For 1liter final mix, dissolve each compound in the volume of water indicated.
The EDTA should be dissolved in boiling water, and the FeSO4 should be prepared last to avoid oxidation.
compound amount water
EDTA disodium salt 50 g 250 ml
ZnSO4.7 H2O 22 g 100 ml
H3BO3 11.4 g 200 ml
MnCl2.4 H2O 5.06 g 50 ml
CoCl2. 6 H2O 1.61 g 50 ml
CuSO4.5 H2O 1.57 g 50 ml
(NH4)6Mo7O24. 4 H2O 1.10 g 50 ml
FeSO4. 7 H2O 4.99 g 50 ml
Mix all solutions except EDTA. Bring to boil, then add EDTA solution. The mixture should turn green. When
everything is dissolved, cool to 70 degrees C. Keeping temperature at 70, add 85 ml hot 20% KOH solution (20 grams / 100 ml final volume).
Do NOT use NaOH to adjust the pH.
Bring the final solution to 1liter total volume. It should be clear green initially. Stopper the flask with a cotton plug and let it stand for 1-2 weeks, shaking it once a day. The solution should eventually turn purple and leave a rust-brown precipitate, which can be removed by filtering through two layers of Whatman#1 filter paper, repeating the filtration if necessary until the solution is clear. Store refrigerated or frozen convenient aliquots. Some people shorten the time for formation of the precipiate by bubbling the solution with filtered air. If no precipitate forms, the solution is still usable. However, you might want to check the pH in this case and adjust it to around 7.0 using either KOH or HCl as needed.
To prepare sulfur-free trace elements for hydrogen generation, the sulfate salts can be replaced with equimolar chloride salts (ZnCl2 10.0 g; CuCl2.2 H2O 1.00 g; FeCl2.4 H2O, 3.60 g).
Reference:http://www.chlamy.org/trace.html
1.Take 50-100mg tissue stored in -80℃ or liquid nitrogen. Grind the tissue into powder in mortar with liquidnitrogen. Transfer the powder into a 1.5ml EP tube(RNase-Free).
2.Add 500μl Trizol, vortex to mix thoroughly and let the tube sit at roomtemperature for 10 min.
3.Centrifuge at 12,000 x g for 10 min at 4℃.Transfer the supernatant into a new 1.5mlEP tube(RNase-Free).
4.Add 100μl chloroform, vigorously shake with hand for 15s and then letthe tube sit at room temperature for 10 min.
5.Centrifuge at 12,000 x g for 15 min at 4℃. Transfer the upper colourless aqueousphase into a new 1.5ml EP tube(RNase-Free).
6.Add 250μl isopropanol, turn the tube upside down many times for around10 min, and let it sit at room temperature for 10 min until the white flocculearise,and then centrifuge at 12,000 x g for 10 min at 4℃.
7.Discard the supernatant,add 500μl,75%ethanol with DEPC,and mixthoroughly.Centrifuge at 7,500 x g for 5 min at 4℃ and discard the supernatant.
8.Dry it at room temperature or in the bechtop for 15 min.
9.Add 15-20μl RNase-Free water to dissolve RNA when the precipitate becometransparent.
10.Store RNA at -80℃.
Protocol of Mosquitoes (Larvae) Feeding
Attention: All team members just can feeding the larvae of mosquitoes. Please inform the administrator before emergence to ensure safe.
This protocol describes mosquito rearing in the insectary. The insectary rooms are maintained at 28°C and ~80% humidity, with a 12 hr. day/night cycle. For this procedure, you'll need mosquito cages, 10% sterile sucrose solution, paper towels, beaker, what man filter paper, glass feeders, human blood and serum, water bath, parafilm, distilled water, clean plastic trays, mosquito food (described below), mosquito net to cover the trays, vacuum, and a collection chamber to collect adults.
Protocol
Mosquito food:
Food A: grounded fish food (Aquaricare). A small pinch needs to be added.
Food B: grounded CAT food (Purina). A small pinch needs to be added.
Food C: cat food (Purina). Two tablets need to be added.
Blood: Animal or human blood can be used to rear mosquitoes. Fresh blood is collected with a syringe and put in a sterile room temperature at 2000 rpm for 5 min. The supernatant is discarded, taking care to remove the buffy coat which comprises other blood cells (e.g. WBC). The RBC pellet is then suspended in an equal volume of RPMI medium by pipetting, and further washed in this medium three times. After the final wash, the pelleted RBCs are resuspended in an equal volume of RPMI medium and stored at 4°C (can be kept for 8-10 days). Just before feeding, the RBC (in RPMI) is centrifuged (2000 rpm for 5 min) to pellet down the cells, and the packed RBC is resuspended in serum (O+ human serum) to obtain a 40% haematocrit. The blood must be kept at 37°C always for the feeding.
DAY 1:
The 3-5 day old adult female mosquitoes are fed on blood to lay eggs.
For blood feeding, an artificial membrane (parafilm) feeding method is used as follows:
Red blood cells (see above for blood preparation) are mixed with heat inactivated serum to obtain a ~40% haematocrit (packed RBC 40% and serum 60%). This is added to the glass feeder.
The feeder is connected to a warm water-jacket (37°C), and placed on the cage to allow the mosquitoes access to the membrane surface.
The mosquitoes are allowed to feed for ~30 minutes.
DAY 3:
The females will lay eggs two days after they feed on blood. A small filter paper wrapped in a conical shape is put in a small beaker containing distilled water, making sure that filter paper gets moist. The beaker is kept inside the cage overnight for the mosquitoes to lay eggs.
DAY 4:
The filter paper containing the mosquito eggs is placed in a plastic tray with ~300 ml distilled water. A pinch of food A is added to the tray and eggs are allowed to hatch to larvae during the next days.
DAYS 5 - 8:
Growing larvae are fed every day with two tablets of food C, and monitored for density and population. On the eighth day (5 day old larvae), the larvae population is diluted from 1 tray to ~10 trays, with a pinch of food A and two tablets of food C in each tray. (~30 mins)
DAYS 9 - 12:
The larvae are fed every day with food C. On the 12th day (9 day old larvae), the water is changed with fresh water, and food is added (a pinch of food B and two tablets of food C). The pupae start developing at this stage. The trays are covered with nets to avoid escape of adults. (~ 8-10 mins)
DAY 13 - 15:
The pupae are allowed to emerge to adults for the next 2 - 3 days. Food is given every day to the larvae/pupae by carefully removing the net to avoid escaping of adults.
DAY 16:
The adults are collected into a cage through an aspirator connected to vacuum. The cage consists of a small 100 ml bottle with a cotton wick that is soaked with 10% sucrose (autoclaved) and a paper towel lining on the bottom to soak any potential sugar spills which may occur during cage handling. (~ 30-40 mins)
DAY 17 - 21:
The adults (both males and females) are then kept in the insectary room for 4-5 days, fed on 10% sucrose before they are again blood-fed to begin the next cycle. The same mosquitoes can be used to lay eggs more than once.
The mosquitoes that are not needed for experiments or rearing can be killed by placing the cage in a freezer. The used trays and cages need to be cleaned and dried before they can be used again.
NOTE: There can be some variations in the mosquito rearing method and different labs may have different techniques.
Reference
Das, S., Garver, L., & Dimopoulos, G. (2007). Protocol for Mosquito Rearing (A. gambiae). Journal of Visualized Experiments: JoVE, (5), 221. Advance online publication. http://doi.org/10.3791/221