</div> </div>

Background |
Pathway |
Optimization |
Results |
|---|
Background
The Modeling portion of the UMaryland iGEM project proposes specific adjustments to the expression levels of enzymes in the methane digestion pathways. Our modeling efforts focus on ensuring the viability of our engineered organism, and optimizing the efficiencies of our pathways. Along the way, our team encountered stumbling blocks that we would like to illuminate for iGEM teams in the future. Incorporated into our modeling page is a concise guide on getting started with the Matlab applet, Simbiology, to model simple metabolic pathways. If you have no interest in this guide, please feel free to optimize your time by skipping over the purple text.
Simbiology is a tool that enables teams with variable modeling backgrounds to build pathway architectures by using a simple drag and drop interface. Although Simbiology is an extremely useful and intuitive tool, it does require some experience to navigate, and it does have its errors; but more on that later. The first step is to open Simbiology by typing “simbiology” into the Matlab command window, then hitting enter.
 Simbiology allows you to drag these elements onto an interface to design your model. For our model, we mainly used the “species” icon and the “reaction” icon.
Simbiology allows you to drag these elements onto an interface to design your model. For our model, we mainly used the “species” icon and the “reaction” icon.
To begin drawing your metabolic pathway, drag and drop a “reaction” icon, located on the left hand sign of the window, onto the blank canvas covering the right half of the window. Then double click on the small orange reaction circle that you placed, and enter your chemical reaction into the block property editor by replacing “null -> null” with your balanced formula. Enzymes are included in the balanced formula. Make sure your enzyme is represented as both a reactant and a product. Clicking out of the Block Property Editor will create species with arrows pointing into your reaction and arrows pointing out of your reaction. Enzymes are connected to the reaction by a dotted line, representing an intermediate that is not consumed or produced by the reaction. You may place as many reactions and species onto the canvas as you would like. If you would like an example model architecture, our pathways are shown later.
Creating the Pathway Architecture
The models for our formate and fructose pathways each consist of two compartments, four reactions and 14 or 15 species, respectively. The initial goal for our project was to produce a co-culture of cells consisting of “sMMO Cells” that produce methanol and “Formate/Fructose Cells” that detoxify methanol, all in an effort to completely biodegrade methane. The first compartment in our model shown below is the “sMMO Cell” that converts methane to methanol, which travels to the second compartment of our model, either the “Formate” or “Fructose Cell.” Each species represents a different molecule involved in our pathway. The architecture for our Formate pathway displays a progression of compounds starting at methane and ending at carbon dioxide. In all, this pathway is facilitated by four enzymes.
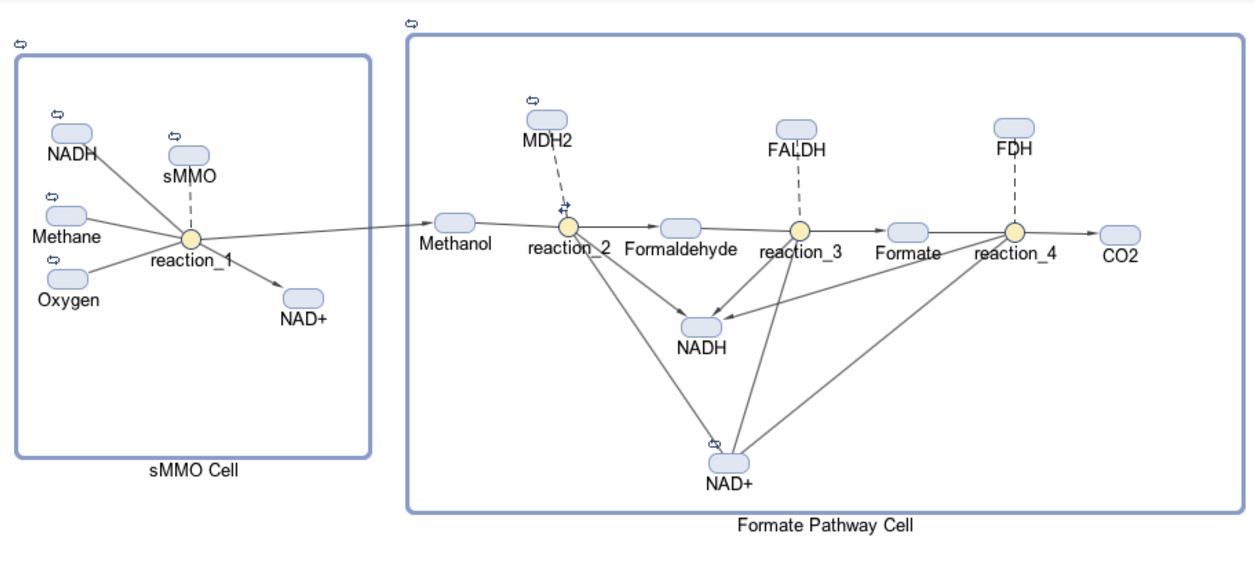
The first half of each pathway is identical. The first enzyme used in each pathway is sMMO, which catalyses the oxidation of methane to methanol while simultaneously oxidizing NADH to NAD+. Both pathways then proceed to oxidize methanol to formaldehyde, which is a reaction catalyzed by the MDH2 enzyme.

Once formaldehyde is produced, the two pathways begin to differ. In the Formate Pathway, formaldehyde is first oxidized to formate, which is then oxidized to carbon dioxide by FALDH and FDH enzymes respectively. In the Fructose Pathway, formaldehyde is incorporated into a 5 carbon sugar by HPS to make D-arabinose 6-phosphate, a 6 carbon sugar. PHI then converts D-arabinose 6-phosphate into D-fructose-6-phosphate, which is a substrate of glycolysis.
The enzyme kinetics for each of the aforementioned proteins are listed in the table below:
| Enzyme | Km | Vmax |
|---|---|---|
| sMMO1 | 3 μM | 56 nanomoles product/minute/mg enzyme |
| MDH22 | 360 mM | 0.09 micromoles product/minute/mg enzyme |
| FALDH3 | 0.09 mM | 5.9 micromoles product/minute/mg enzyme |
| FDH4 | 1400.0 mM formaldehyde34.0 NAD+ | 0.625 micromoles product/minute/mg enzyme |
| HPS5 | 0.74 mM formaldehyde0.081 mM D-ribulose-5-phosphate | 5.9 micromoles product/minute/mg enzyme |
| PHI6 | 0.029 mM | 20 micromoles product/minute/mg enzyme |
Enzyme kinetics can be entered into the Reaction Editor by double clicking on the small orange circle representing a reaction. First choose a Kinetic Law, then enter the kinetic values simbiology asks for. Simbiology will then use the Kinetic Law and kinetic values to produce a reaction rate.
The standard units for Vmax are micromol product/minute/milligram enzyme, however, Simbiology initially asks for Vmax in terms of micromole/minute. To the best of our knowledge the easiest way to include enzyme amount as a factor in your model is by manually changing the units of Vmax to micromole/minute/micromole by clicking on the unit drop down menu and selecting “Create new unit” (Vmax must be represented in units of micromole/minute/micromole because of dimensional analysis constraints in Matlab, but keep in mind that you will most likely enter enzyme amount as a mass value). After changing Vmax units, you must change the ReactionRate by multiplying the given formula by the name of your enzyme. At any point in the creation of your model, you may double click on the species representing your enzyme and enter an amount value (leave the enzyme amount in terms of micromole).

This stage of creating our Simbiology model presented a subtle, yet extremely significant stumbling block to our progress. The drop down menu for units allows one to select different prefixes for units. Unfortunately, it seems as though these prefixes were not properly integrated into the software, and Simbiology will not perform the necessary dimensional analyses to convert between micro and milli, for example. Therefore, it is crucial that all your analogous units employ identical units. For example, our modeling team decided to work exclusively with a “micro” prefix for concentration and molarity units. 56 nanomoles became 0.056 micromoles and 300 millimolarity became 300,000 micromolarity.
This leads us to an important piece of advice: do not implicitly trust everything the program claims to do. Our modeling team discovered this discrepancy in the Simbiology software by running models in parallel with certain hand done calculations. When our model did not remotely resemble our paper and pencil calculations, we knew there was an error in the way Simbiology handled units. We recommend that in addition to creating your desired model, you create a control model where the outcome is known, or perform hand-written calculations to check certain parts of your model.
From this point on, Simbiology runs fairly smoothly, so we’ll leave it to you to figure out the rest. Best of luck!
After we created the basic architecture for our two models, we experimented with various parameters to optimize our pathway.




