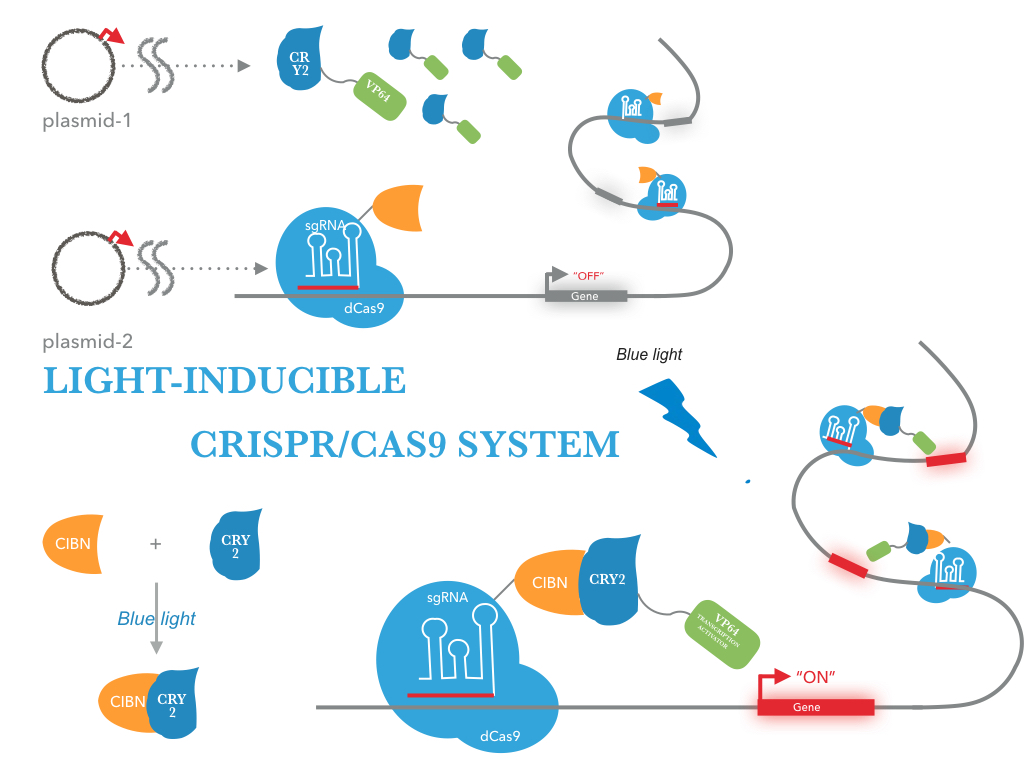
In this work, we aim to develop a light inducible CRISPR/Cas9-mediated gene activation system in prokaryotic and lower eukaryotic cells, to precisely manipulate target gene activation upon the light induction. We fused two light-activated plant proteins, CRY2 or CIBN, to a transcriptional activator VP64 or the catalytic domain deleted tCas9, generating CRY2-VP64 or tCas9-CIBN chimeric constructs. In the presence of a specific sgRNA guide, the tCas9 domain from the tCas9-CIBN binds to the promoter sequence of a reporter gene. Once exposing to blue light, the CRY2 domain from the CRY2-VP64 can form a protein complex with tCas9-CIBN, which in turn brings the VP64 domain to the chromatin and activates the reporter gene. We have successfully generated a prokaryotic cell strain that the activation of reporter gene is regulated by blue light.
Light-inducible CRISPR/Cas9 system
The NEU-China iGEM team sought to create a new mode of gene editing and manipulation for use in prokaryotic and lower eukaryotic cells. We generated two expression systems harboring CRY2-VP64 and tCas9-CIBN chimeric constructs. The introduction of a specific sgRNA, the tCas9-CIBN protein binds to the promoter region of the reporter gene (GFP) through tCas9-sgRNA complex, without the activation of reporter gene. In the presence of blue light, the CRY2-VP64 forms a complex with tCas9-CIBN, via CRY2 and CIBN protein-protein interaction. The expression of the reporter gene is then activated by VP64.
Over the summer, we accomplished the following:
Getting CRY2 and CIB1 gene from Arabidopsis thaliana
Total RNA was isolated from Arabidopsis thaliana and mRNA was reverse transcribed into cDNA. CRY2 and CIB1 gene was amplified using primers.
Creating/Testing prokaryotic tCas9-CIBN chimeric constructs
We constructed tCas9-CIBN chimeric constructs gene with an prokaryotic RBS sequence from the Community collection (BBa_B0034). For detection of expression the fusion protein was tagged with a HA-epitope coding sequence (BBa_K1150016).
Creating/Testing prokaryotic CRY2-VP64 chimeric constructs
We constructed CRY2-VP64 chimeric constructs gene with an prokaryotic RBS sequence from the Community collection (BBa_B0034). For detection of expression the fusion protein was tagged with a FLAG-epitope coding sequence (gactacaaggacgacgacgacaaa).
Creating/Testing prokaryotic tCas9-VP64 chimeric constructs
To address these limitations, we adapted the CRISPR/Cas9 activator system for optogenetic control. We constructed tCas9-VP64 chimeric constructs gene with an prokaryotic RBS sequence from the Community collection (BBa_B0034).For detection of expression the fusion protein was tagged with a FLAG-epitope coding sequence (gactacaaggacgacgacgacaaa).
Testing tCas9-VP64
We tested our tCas9-VP64 fusion protein by transfecting it into BL21 cells along with a guide RNA which will target the tCas9-VP64 to upstream regulatory sites on our reporter construct and activate the expression of GFP. We showed that our tCas9-VP64 fusion is fully functional.
Verification of the suppression efficiency of gRNAs
To ensure the suppression efficiency of the gRNA, four gRNA sequences targeting different sites of CSPA promoter were designed and transformed into the E. coli strains BL21. Efficient suppression of CSPA promoter in strains BL21 was observed. The GFP level dramatically decreases after transforming a fragment that expresses CSPA promoter gRNA. The sequence with the best suppression effect was selected for further study.
Silencing capability validation
We next evaluated the effect of tCas9-CIBN on suppressing CSPA promoter. The GFP expression level was assayed in strains BL21 after co-transformation with tCas9-CIBN and gRNA in the absence of light conditions. Compared with control groups, green fluorescence intensity and mRNA levels were dramatically reduced in groups treated with gRNA and tCas9-CIBN.These results suggest that gRNA can specifically guide tCas9 to target upstream of CSPA promoter, thereby inhibiting CSPA promoter to reduce GFP expression levels.
Testing tCas9-VP64
We tested our tCas9-VP64 fusion protein by transforming it into BL21 cells along with a guide RNA which will target the tCas9-VP64 to upstream regulatory sites on our reporter construct and activate the expression of GFP. We showed that our tCas9-VP64 fusion is fully functional.
Application:Light-inducible Gene Activation
We construct this new system in prokaryotic cells and to explore the optimal combination of these two techniques in order to enable the most efficient gene regulation with light. We have generated a GFP expression system that is blue light inducible in E. coli. The activation of the GFP gene would require targeting an activator (vp64) to a sequence upstream of the promoter of GFP. tCas9 allows us to target the promoter of GFP using a guide RNA specific to that sequence.
We also Improved a previous iGEM project
We improved CRY2 (BBa_K1592015) and CIB1 (BBa_K1592016). These biobricks are key BioBrics in " A live eukaryotic cell based auto-cementation kit " project of HUST-China 2015. CIB1 encodes a basic helix-loop-helix (bHLH) protein, would interact with cryptochrome 2 (CRY2), a blue light stimulated photoreceptor. This fusion protein is for use in a yeast-two-hybrid system, and a Gal4 DNA activating domain fused to N terminus of CIB1. To regulate DNA transcription by blue light, the system is based on a two-hybrid interaction in which a light-mediated protein interaction brings together two halves (a binding domain and an activation domain) of a split transcription factor.
We improved the function of CRY2 (BBa_K1592015) and CIB1 (BBa_K1592016) and enter our parts (BBa_K1982005 and BBa_K1982000) in the Registry. Please see the Registry page of this two parts.



















