Our Project

The concept of BioBricks was first introduced by Tom Knight at MIT in 2003. His vision was to standardise synthetic biological parts in a similar way to Lego bricks that interlock to form larger constructs. In turn, this has enabled research groups to engineer novel biological systems. Our team has been inspired by Tom Knight’s “Lego-like” approach to synthetic biology.
However, we wondered how we could mix electronic devices with synthetic biological devices. Our project therefore involves us replacing some of the traditional electronic components in a circuit with biological alternatives. Like Lego, the circuit will allow synthetic biologists to mix and match bacterial and electronic components to create electro-biological circuits. This approach will represent a foundational advance in the way synthetic biological circuits are designed and implemented.
We believe that by merging biology, electronic engineering and computer science, our project also holds fantastic opportunities for education. There is a common misconception that all bugs are bad, but our project will demonstrate the benefits of bacteria in a fun and safe manner.
Synthetic biology is an exciting, and unique field that we want to make accessible to children worldwide. We want the next generation to have a better understanding of what synthetic biology is, and inspire new ways to apply synthetic biology to real world applications. Perhaps our circuits will alter the perception of Genetically Modified Organisms, inspire a new way to generate sustainable electricity, or even work towards creating a computer made entirely from synthetic organisms.
Design of Hardware

Having decided on an electrical theme, we were inspired by educational electronics kits to build a standardised breadboard-style testing system for our constructs that could also be used for demonstrations. This was a great chance to reach out to the local community for help and advice, as Newcastle has its own local maker group We would go on to work with lasercutters and 3D printers to produce parts to fulfil our own specifications.
Initially we worked with FabLab, a local fabrication workshop based in Sunderland to come up with our initial designs, building prototypes and investigating the best ways to join parts together. Later we moved our base of operations to OpenLab within Newcastle University where we were assigned a design intern who helped us bring our final designs to life. We opted for a system that used magnets which allow the user to freely interchange parts and pass electricity through our microfluidic devices.
This was a fantastic learning opportunity for us, as none of us had any previous design experience, and you can read more about our design process here
Design of Parts

Our goal of creating biological analogues of electronic devices offered us a huge range of potential biobricks. At the planning stage, we designed theoretical versions of many standard components, including lightbulbs, batteries, variable resistors and even a capacitor!
Our parts rely on electrical heating for activation, allowing integration into existing electrical systems. We have created functional versions of our lightbulb and variable resistor designs, along with an improved efficiency microbial fuel cell based on a design proposed by Team Bielefield in 2013. We also developed a simulator that predicts the output of our devices, whether alone or integrated into more complex systems.
Being able to work on such a range of different devices has allowed us to learn a great deal about cellular mechanisms, enhancing our general biological knowledge. It was really exciting to try and combine the two different disciplines of electrical and genetic engineering.
Design of Human Practices

As we're competing in the Foundational Advance track we felt obligated to consider, at great length, the ethical issues that could be raised by future development in the new field of science we're striving to lay the foundation for. After discussing with experts from PEALS (The Policy, Ethics and Life Sciences Research Centre at our University) it became apparent that due to the wide range of potential applications our research could contribute towards - our end Human Practices output had to take a holistic view.
After considering this and looking at the strengths of our team, we decided to utilize the skills of the three computer scientists on the team and program a simulator to act as a thought piece on the future implications of Bio-electrics. The simulator features 5 levels which provide different scenarios, each raising potential ethical concerns. Each level carries an inspiration from our interactions with others - whether it be our discussions with PEALS, talking to a space expert who inspired the the Mars Level or the sixth formers day we ran, from which one student's experiences with a Kidney dialysis machine forms the narrative behind another level.
You can read more about our design process for Human Practices here
Modelling
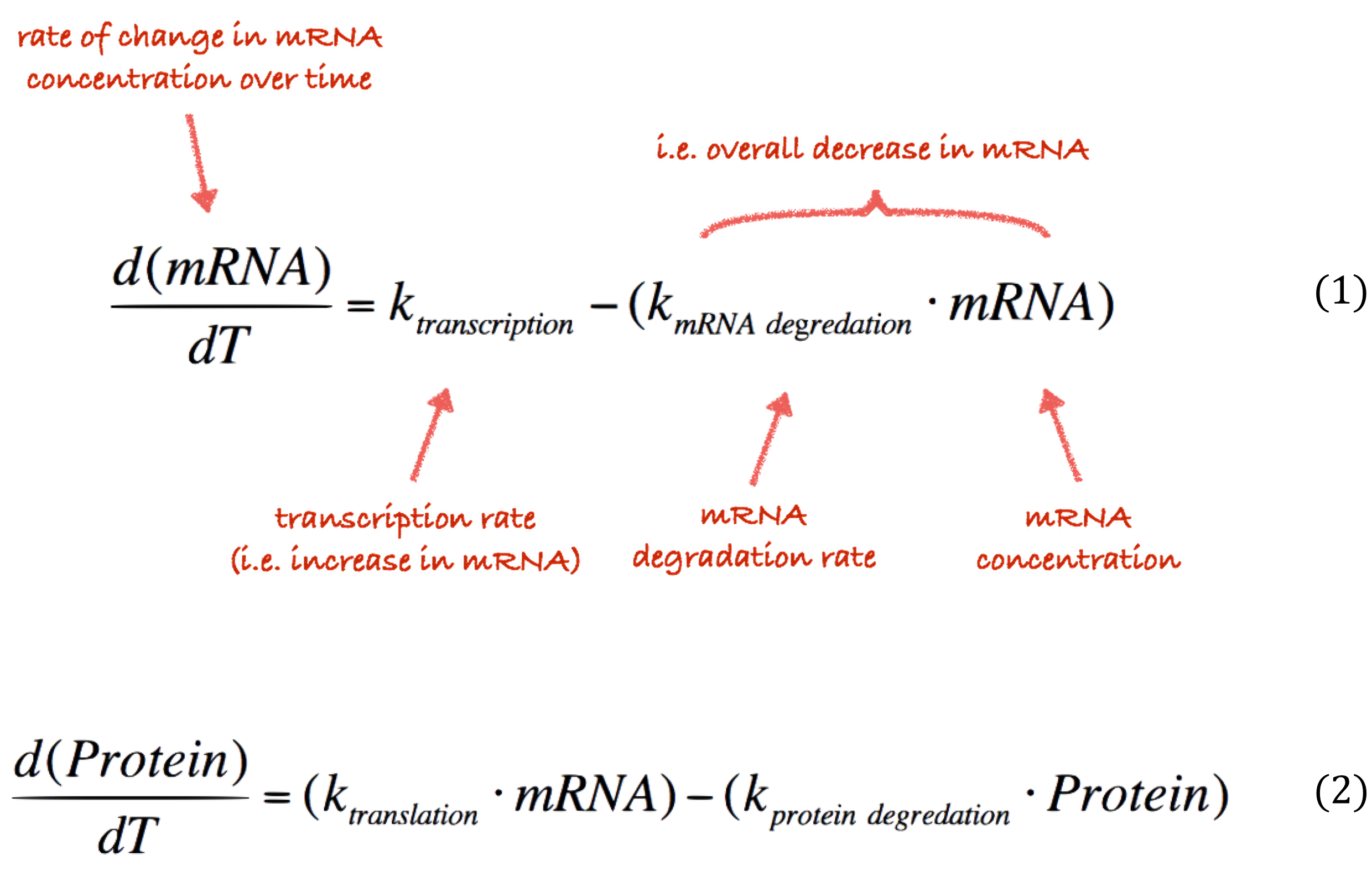
Our teams goal is to show how our technology could work in practice. Consequently, we used modelling to explore potential uses of our technology beyond the summer, and to inform our design process. We used a number of different modelling tools, both for designing our biological constructs as well as our physical hardware. For our phsyical hardware we made use of the COMSOL software to perform multiphysics modelling whist for the biological constructs we usd RuleBender and rule based modelling as well as our own ordinary differential equation modelling.
All of these gave us valuable insights into our design process. For instance, after we conducted multiphysics modelling of our original chamber designs we found they were too large. Our modelling showed us that we needed to switch to microfluidics scale hardware. Modelling also gave us new directions to explore, for example during the modelling of our resistor constructs we came upon the idea of a cell-free system. Finally, modelling gives us a way to showcase how bacteria and electronics might be integrated through our ‘simulator’ which models our constructs in circuits users can build themselves.
Results

Over the course of the summer, we have performed a large number of experiments, both to inform further research and to help with characterising our parts. The earlier tests were vital in directing the flow of project, indicating the level of electrical current we would need and allowing us to refine the design of our breadboard around integration of the microfluidic devices.
Proof of Concept

We intended for our constructs to be used in self-contained microfluidic devices, which will allow quick and sufficient heating produced from an electrical current in order to induce our ‘light bulb’ constructs.
We also wanted to be able to significantly scale down the microbial fuel cell to a size similar to the bulb component in order to be compatible with our breadboard while also producing a measurable voltage.
Demonstration

In order to achieve a gold medal, iGEM teams can demonstrate their constructs working under real world conditions. With this in mind, we have used our breadboard kit hardware to electrically heat transformant bacteria and activate our 'lightbulb' biobricks.
Electrical current is transferred through the combination of magnets and conductive tape in the breadboard, through to the electrodes in the microfluidic device. The test detailed illustrates how GFP production in our 'lightbulb' bacteria can be activated in simulated real-world conditions using standard equipment.
Protocols

We have used a wide variety of protocols throughout our project, including previously designed standards such as the University of Reading's fuel cell preparation guide, alongside those we have written ourselves, for example creation of our microfluidic microbial fuel cells.

