Iosikharenko (Talk | contribs) |
Iosikharenko (Talk | contribs) |
||
| Line 131: | Line 131: | ||
=Result= | =Result= | ||
==Cell Cycle Dependent Promoter: Pnrd== | ==Cell Cycle Dependent Promoter: Pnrd== | ||
| − | As described in the systems page, a cell division dependent promoter is essential for the functioning of our intergenerational phenotypic switching device. Stanford-Brown iGEM 2012 documented the characterisation of promoter Pnrd (BBa_K847211) which activates during the cell division phase and confirmed its functionality. However, since it was submitted in RFC[25] standard, we decided to improve the part by using designing the part in RFC[10] in order to deal with the disadvantages of RFC[25] standard. | + | As described in the [https://2016.igem.org/Team:UT-Tokyo/Project#system systems page], a cell division dependent promoter is essential for the functioning of our intergenerational phenotypic switching device. Stanford-Brown iGEM 2012 documented the characterisation of promoter Pnrd ([http://parts.igem.org/Part:BBa_K847211 BBa_K847211]) which activates during the cell division phase and confirmed its functionality. However, since it was submitted in RFC[25] standard, we decided to improve the part by using designing the part in RFC[10] in order to deal with the disadvantages of RFC[25] standard. |
[[File:T--UT-Tokyo--Model-Result--1.jpg]] | [[File:T--UT-Tokyo--Model-Result--1.jpg]] | ||
Revision as of 23:47, 14 October 2016
Introduction
The researches conducted so far have mostly been transforming Escherichia coli by changing external environments or introducing inductive factors. To the best of our knowledge, phenotypes of E. coli switch without any input from the exterior.
Until now, no study in iGEM competition has been done on changing autonomously phenotypes of E. coli before and after cell division.
Therefore, we have designed a system which automatically generates three phenotypes of E.coli every time a cell divides.
Specifically,in our system, if E. coli in which first generation of gfp is found divides, we cannot find gfp anymore but only rfp. If this second generation of E. coli divides then instead of rfp, cfp is expressed.
After another division in the fourth generation instead of cfp we find gfp again. In this system, the phenotype of E. coli differs from each other between the (3n)th, (3n+1)th, and (3n+2)th generation.
Figure
As we have created this genetic circuit,sigma factor, toehold switch and Pnrd promotors (link to the system) are chosen. In modeling we use them to simulate the system we made .(link to modeling) Modeling shows us whether the genetic circuit builded in thoughts is realizable.
In addition, after confirming the system runs on modeling, we ran an inspection on parts and devices in the experiment.
System
By designing a genetic device that shifts the dominantly expressed protein among GFP, RFP and CFP every cell cycle, our system can realize E. coli that in turn gives out green, red, blue (and green again) fluorescence. To understand how it works, let’s first take a look at the individual components we used. These include Toehold Switch, sigma factors, Pnrd, and PBAD.
Explanatory Notes
Genes are shown in the diagram below, where left represents the upstream.
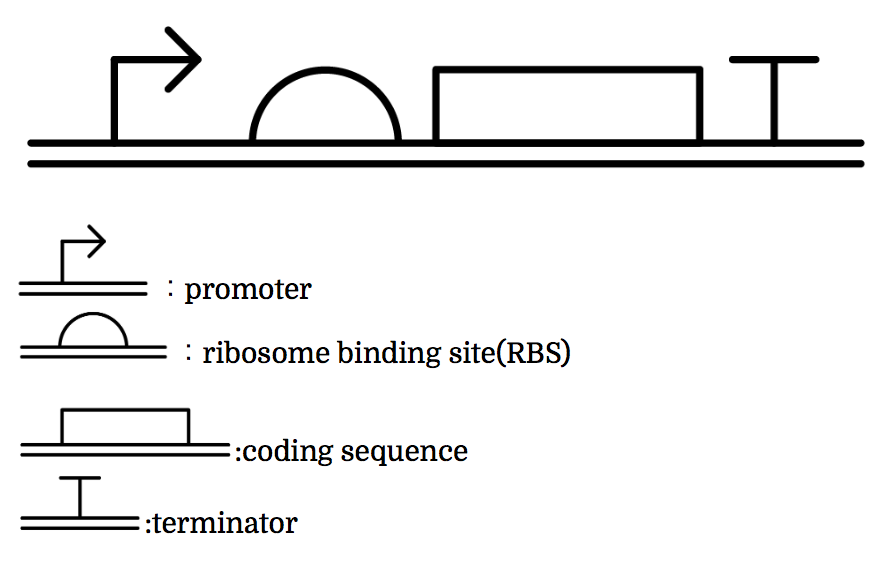
Toehold Switch
Toehold Switch is a translational regulation system which takes advantage of the bonding between specific transcripts. Genes downstream of the Toehold Switch on a mRNA cannot be translated without the existence of specific trigger RNA.
Ribosomes cannot bind to toehold RBS under normal condition. Therefore, proteins cannot be made because mRNAs cannot be translated even when the gene is transcribed. For a ribosome to bind to toehold RBS, the existence of a specific short RNA named trigger RNA is necessary.
The previous gene which the toehold RBS stops transcription comes to the state below under the existence of the trigger RNA, and will be translated. mRNAs being translated are represented as yellow ones.
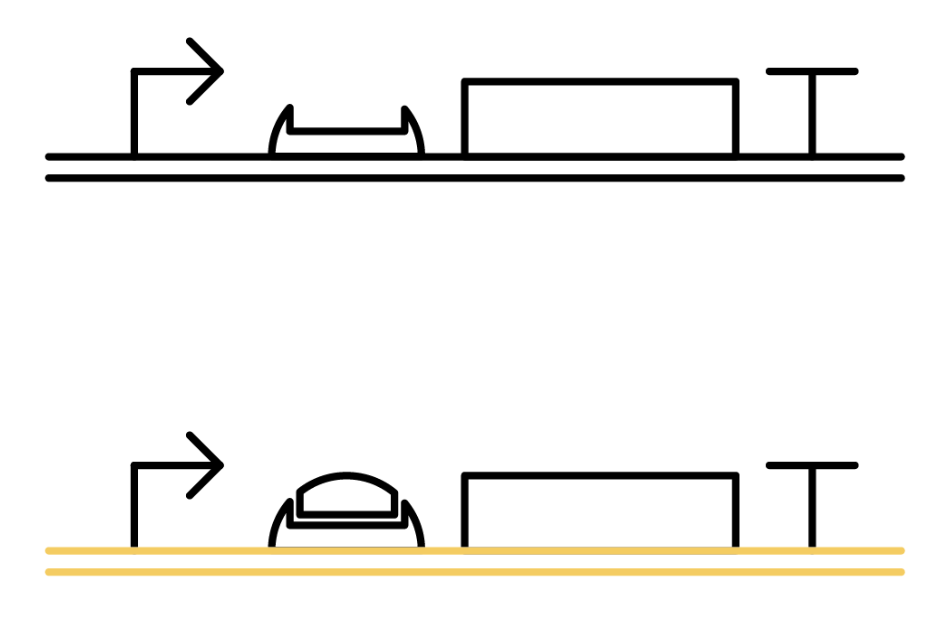
Why ribosome cannot bind to toehold RBS?
Specific sequences, called ribosome binding site (RBS) to which ribosome binds exists on mRNAs . Inserted with specific complementary sequences on both upstream and downstream of RBS, mRNA forms a hairpin structure that encloses the RBS. Ribosome therefore cannot bind to toehold RBS and thus genes downstream unable to be translated. This kind of RNA is called switch RNA. However, the hairpin structure of a switch RNA can be broken by what is called the trigger RNA, which includes partially complementary sequence with the switch RNA and thus makes the translation of downstream genes possible.

Sigma Factor
The promoter below (sigma promoter) works only under the existence of sigma-factor, which binds to and then activates the promoter. However, anti-sigma factors also bind to sigma factors, which competitively inhibit sigma factors from functioning.
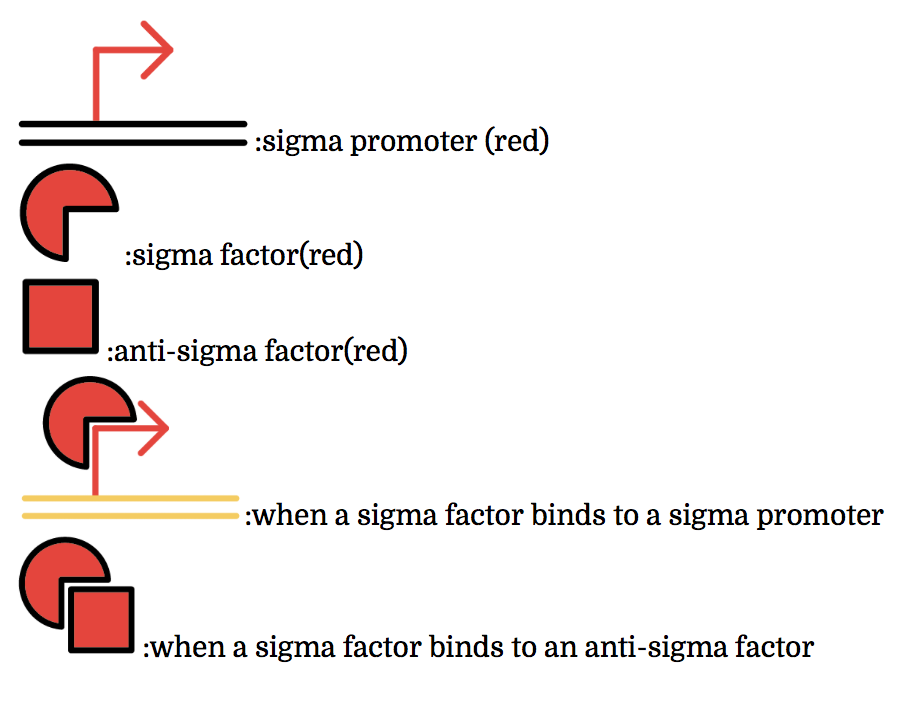
The sigma factors are promoter recognition subunits of RNA polymerase. A sigma factor is associated with a part of promoters. A sigma factor recruits RNA polymerase to its corresponding promoter and initiates transcription. Sigma factors have great variety. Some of sigma factor-promoter pairs have one-to-one correspondence. Thus, if only sigma factors which have one-to-one correspondence are used, a transcription activating system in which a sigma factor activates the transcription only from corresponding promoter can be made.
Anti-sigma factors are proteins which are related to transcriptional control mechanism by sigma factors. An anti-sigma factor inhibits the binding between RNA polymerase and sigma factor. Consequently, anti-sigma factors repress the transcription from the promoters which sigma factors initiate. In the same way as sigma factors, anti-sigma factors have great variety and some anti-sigma factors prevent only a specific sigma factors from transcriptional control. Therefore a transcription control system (i.e. not only activating but also repressing) can be constructed by using specific sigma factors and anti-sigma factors which have one-to-one correspondence.[2](words underlined were grammatically modified from the original)
Multiple combinations of sigma factors, sigma promoters, and anti-sigma factors exist. Every factor we used functions exclusively within its combination and does not bind with factors of other combinations. In other words, they do not crosstalk. To make this easier to understand, factors and promoters of the same combination are presented with the same color.
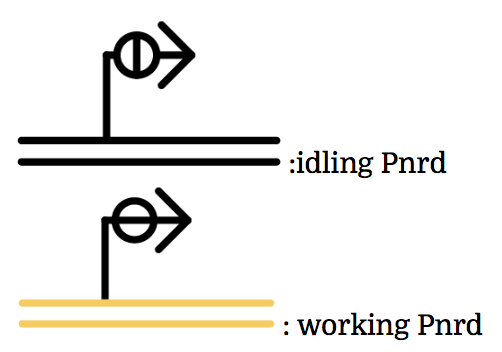
Pnrd
Pnrd, another kind of a promoter, works only during cell division.
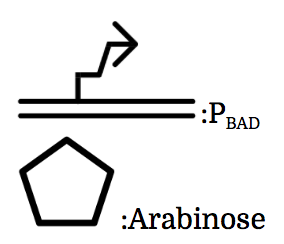
PBAD
We also installed a promoter called PBAD, which works only under the existence of arabinose, drawn as a pentagon in the chart.

Now we’ve seen the 4 components and they will help us understand what our team wants to make. Let’s go to the journey to the gene constructions and systems.
Gene Constructions and Systems
The constructions we want to make are explained below. Compounds such as RNAs and proteins that exist in the cell at a specific time are presented in the yellow frame in the upper right. The 8 genes below are our system to create multiple phenotypes over generations.
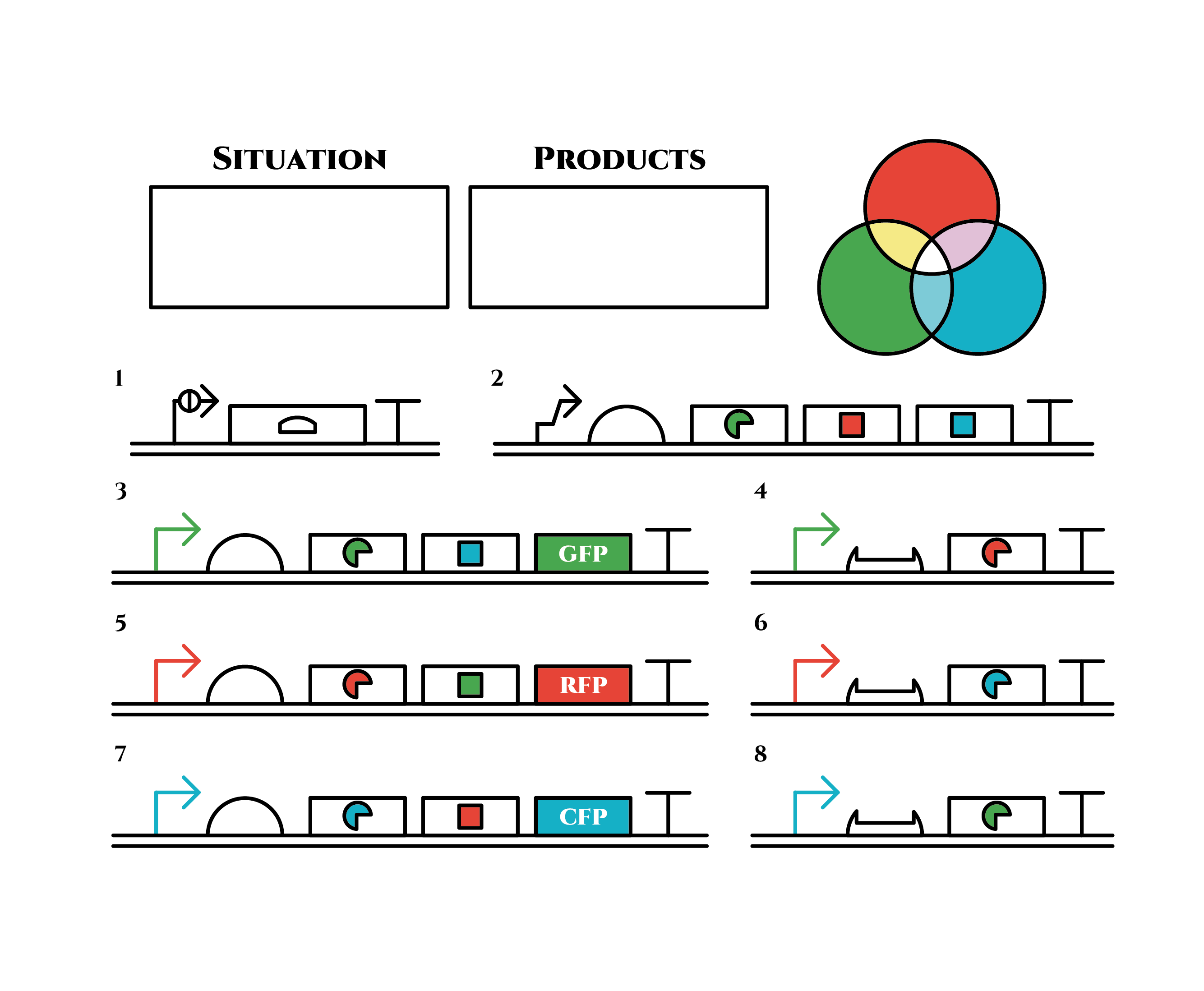 So, how these genes go?
So, how these genes go?
Here is the state before phenotypic change. From the beginning, arabinose is added into the culture medium.
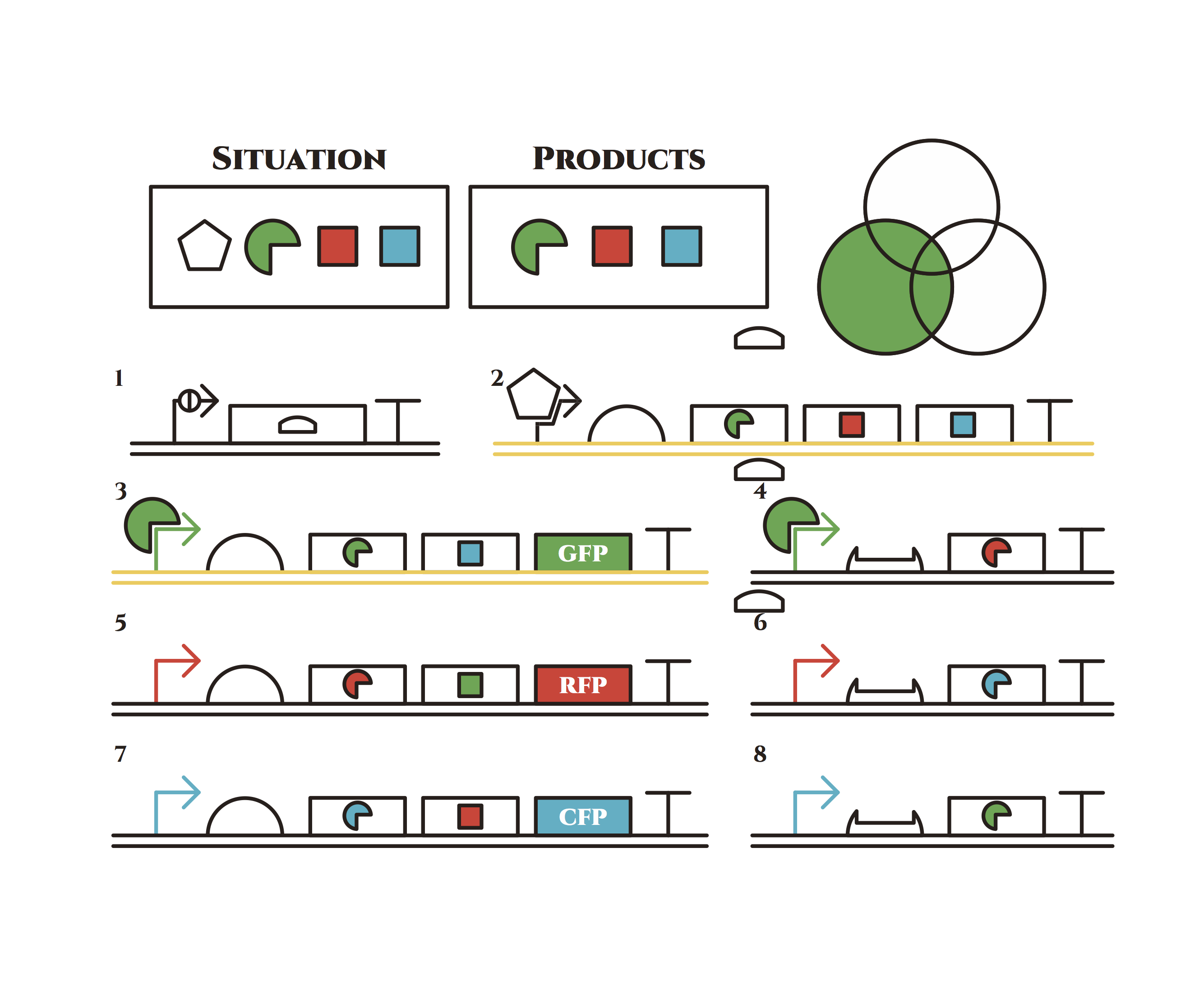 Arabinose induces the expression of the genes in the upper right. As a result, genes in the second row are expressed under a positive feedback. Because of this positive feedback, the genes will be expressed stably without arabinose here on. The mRNAs transcribed from the left gene are translated, while those from the right gene are not because of the lack of trigger RNA.
Arabinose induces the expression of the genes in the upper right. As a result, genes in the second row are expressed under a positive feedback. Because of this positive feedback, the genes will be expressed stably without arabinose here on. The mRNAs transcribed from the left gene are translated, while those from the right gene are not because of the lack of trigger RNA.
Then, we removed arabinose from the culture.
At this time, despite of the degradation of the expressed factors at the upper right, new factors continue to be expressed from the left genes in the 2nd row . Notice the cell is still not dividing at this point.
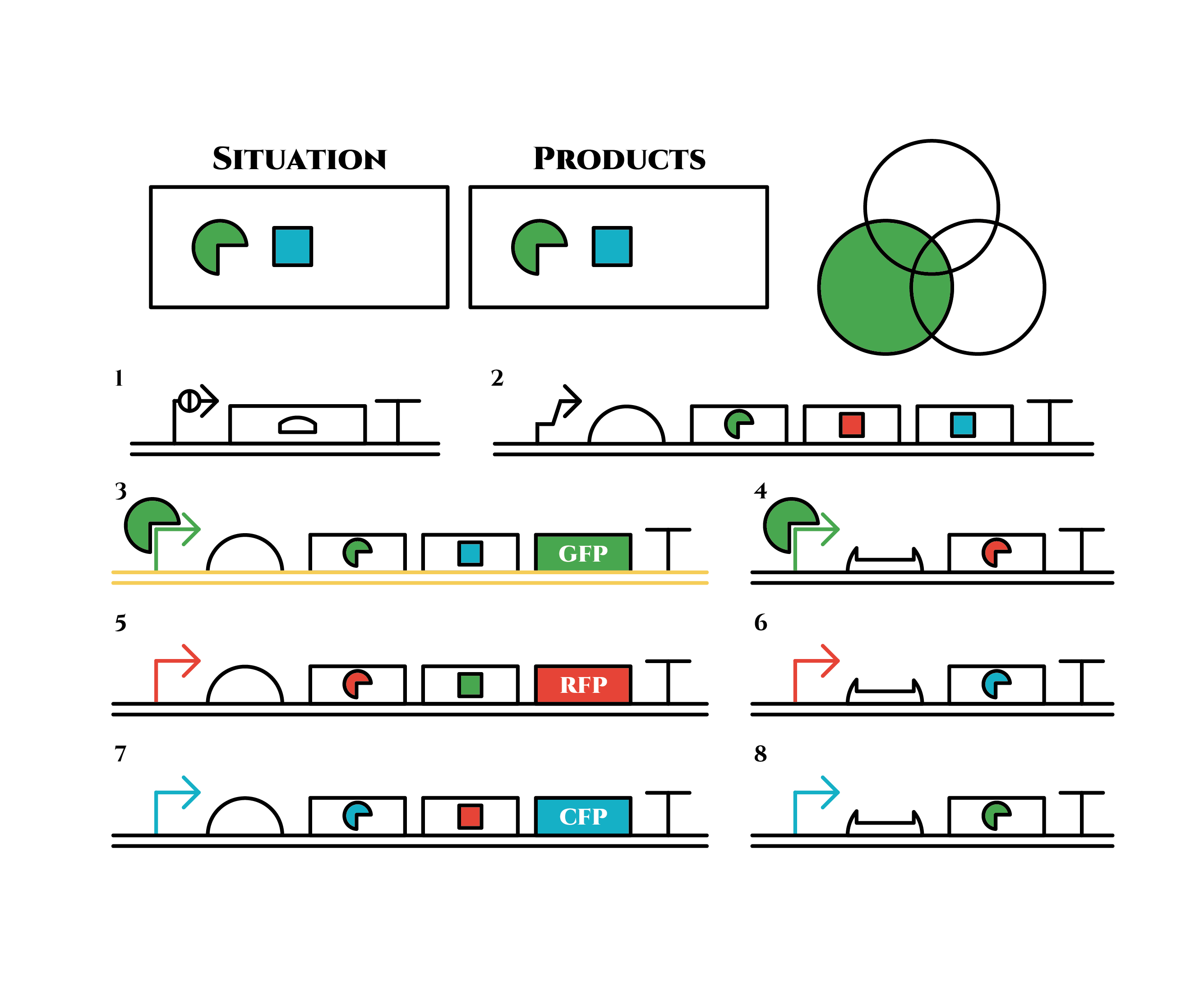 Red sigma factors are made from the left gene on the 2nd row, which in turn bind with the red sigma promoter and make more new red sigma factors. In this way, the gene promotes its own expression and this leads to the stable expression.
Red sigma factors are made from the left gene on the 2nd row, which in turn bind with the red sigma promoter and make more new red sigma factors. In this way, the gene promotes its own expression and this leads to the stable expression.
And what happens when cell division occurs?
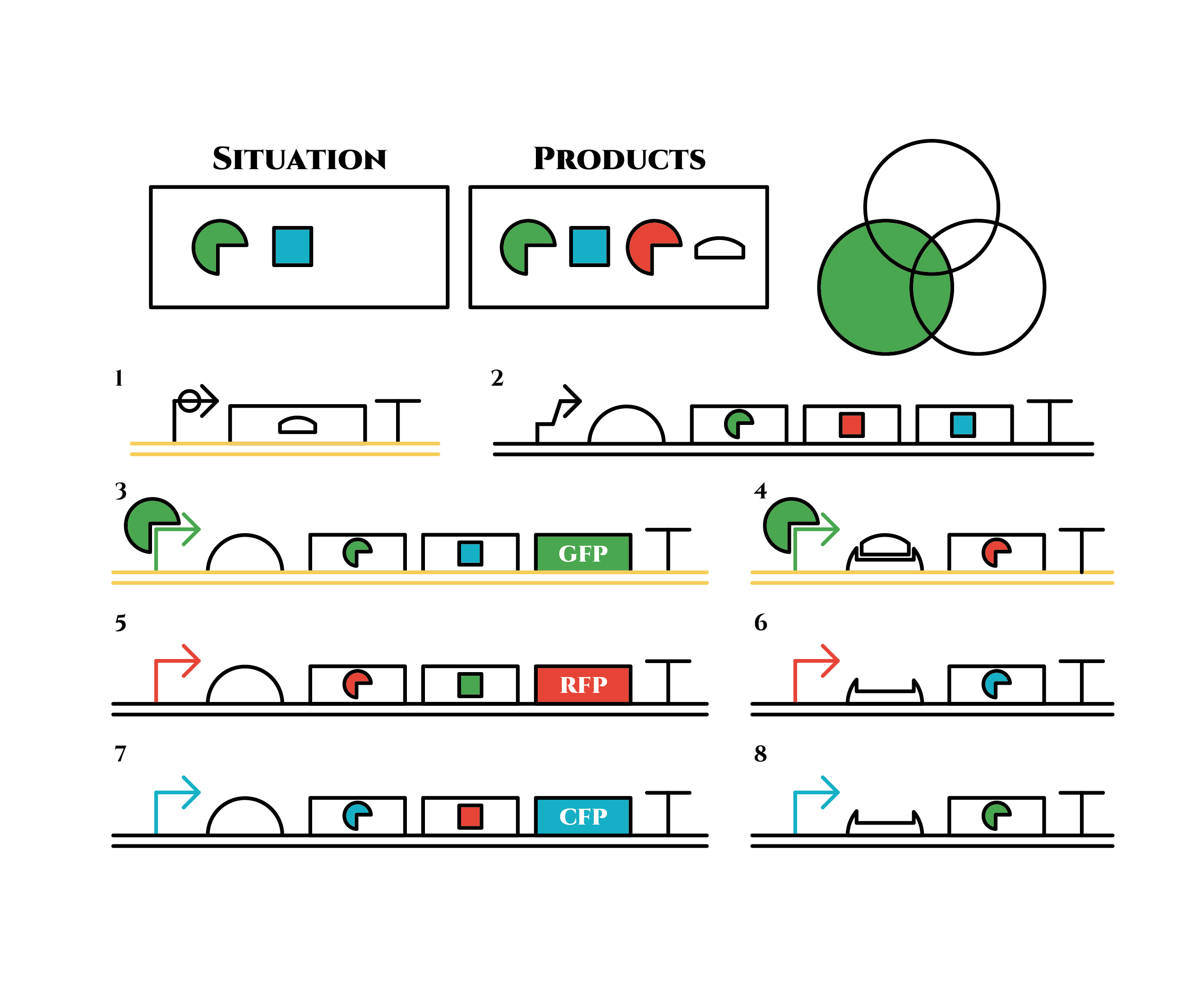 The left gene in the 1st row expresses and trigger RNAs are produced when cell division occurs. Then, let’s focus on the right gene on the 2nd row. Genes downstream can only be expressed with both the existence of a red sigma factor and the trigger RNA, which seem like an AND gate,a kind of basic digital logic gates.
The left gene in the 1st row expresses and trigger RNAs are produced when cell division occurs. Then, let’s focus on the right gene on the 2nd row. Genes downstream can only be expressed with both the existence of a red sigma factor and the trigger RNA, which seem like an AND gate,a kind of basic digital logic gates.
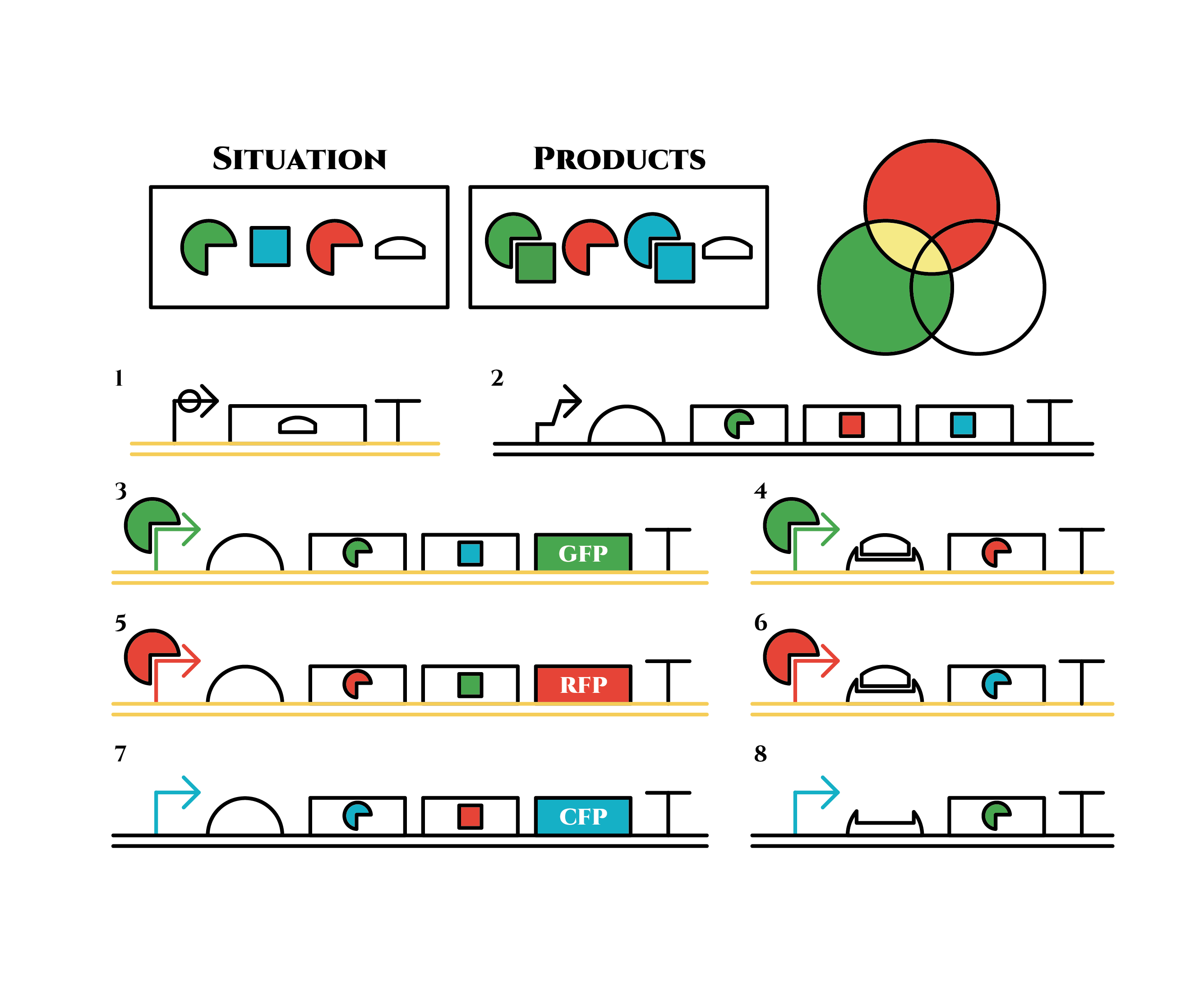
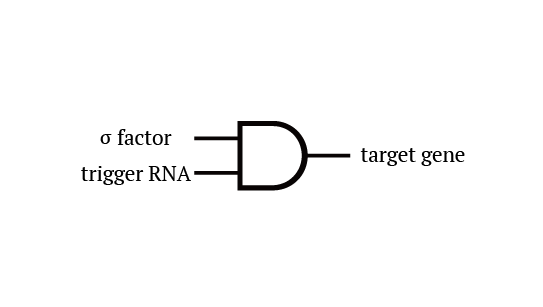 Now we obtained a trigger RNA and a red sigma factor still remains, so the right gene in the 2nd row finally begins to be expressed and blue sigma factors are produced. Then the gene expression will change as below.
Now we obtained a trigger RNA and a red sigma factor still remains, so the right gene in the 2nd row finally begins to be expressed and blue sigma factors are produced. Then the gene expression will change as below.
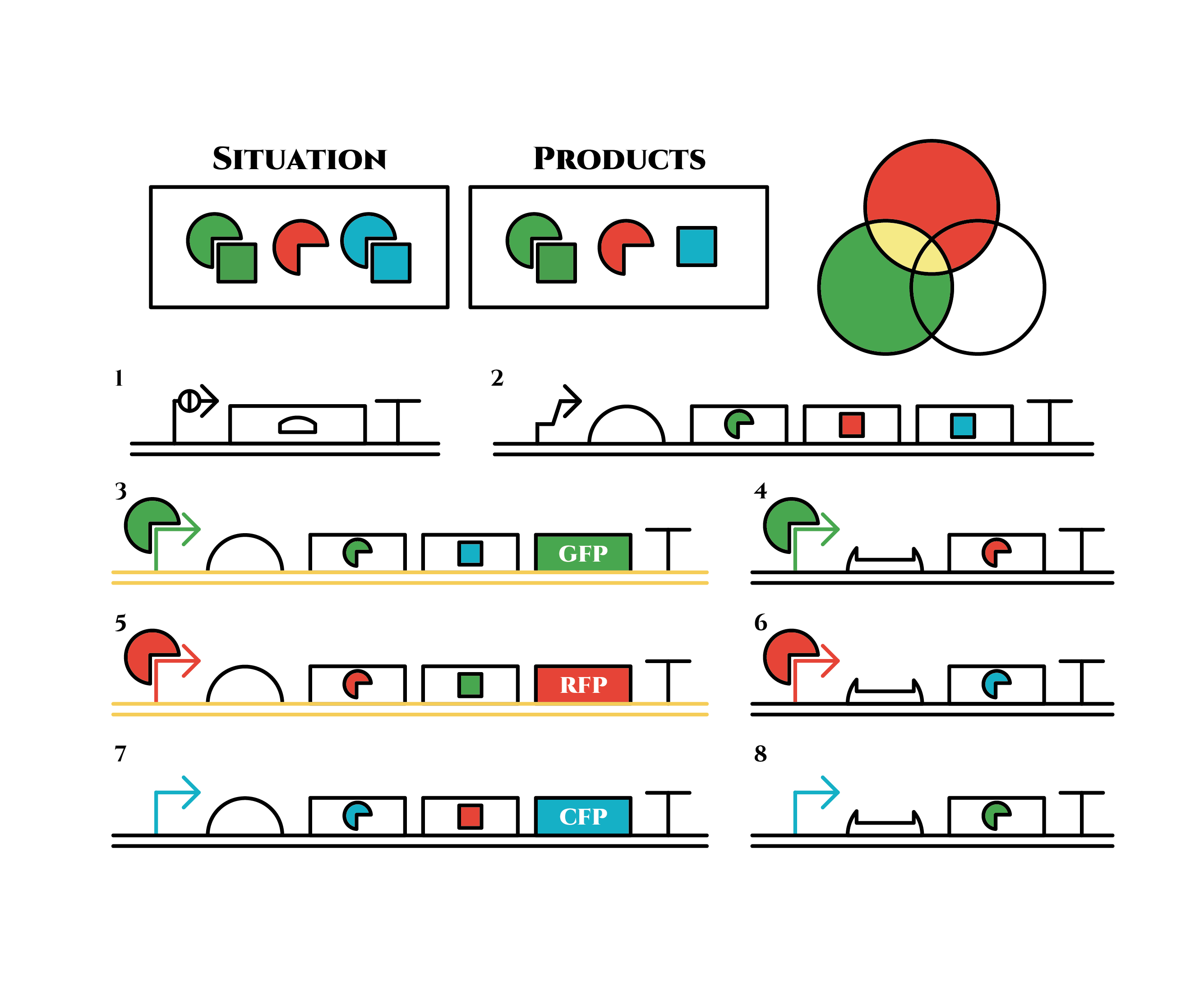
 Under the existence of blue sigma factors, genes in the 3rd row are expressed . Blue sigma factors, red anti-sigma factors, green sigma factors are made. While blue sigma factors work on the blue sigma promoter, newly-made green sigma factors do not affect the green sigma promoter because of its small number against the many green anti-sigma factors that exist already. On the contrary, the small number of newly-made red anti-sigma factors cannot block the already-existing red sigma factors’ effect on the red sigma promoter completely.
Under the existence of blue sigma factors, genes in the 3rd row are expressed . Blue sigma factors, red anti-sigma factors, green sigma factors are made. While blue sigma factors work on the blue sigma promoter, newly-made green sigma factors do not affect the green sigma promoter because of its small number against the many green anti-sigma factors that exist already. On the contrary, the small number of newly-made red anti-sigma factors cannot block the already-existing red sigma factors’ effect on the red sigma promoter completely.
Here is the state after cell division.
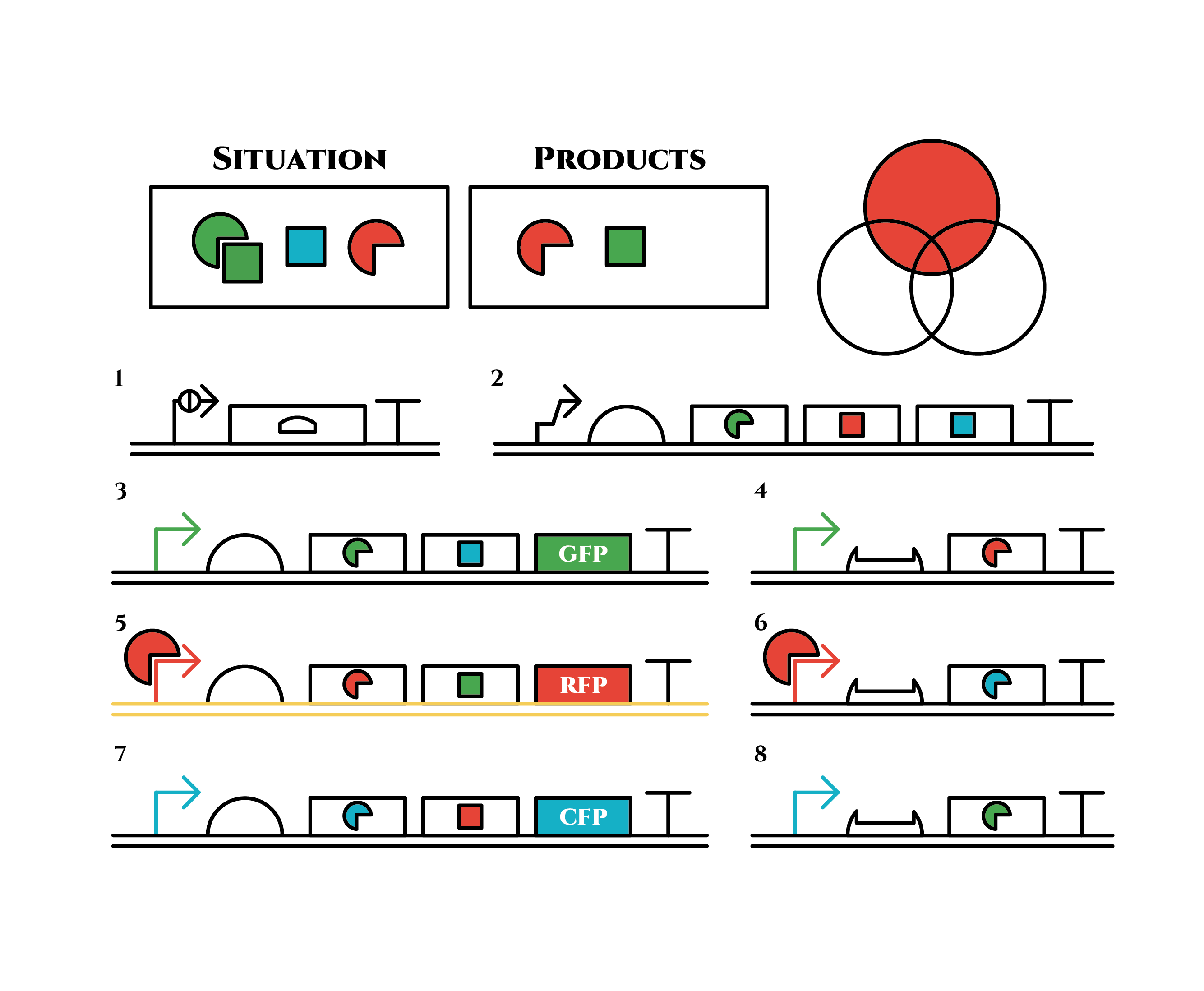
 The genes on the right stop being translated. Plenty of blue sigma factors and red anti-sigma factors are made from the left gene in the 3rd row. The expression of the left gene in the 2nd row gradually declines because of the increase of red anti-sigma factors.
The genes on the right stop being translated. Plenty of blue sigma factors and red anti-sigma factors are made from the left gene in the 3rd row. The expression of the left gene in the 2nd row gradually declines because of the increase of red anti-sigma factors.
When there are sufficient red anti-sigma factors, the gene expression will finally become as following.
 This is the exact same state as the phenotypic changes began, except that it is the genes in the 3rd row, instead of the 2nd, that are being expressed. In other words, the expressed genes are switched through one cell division. The gene groups are symmetric in terms of colors (red, blue and green) when the reagent does not exist. Therefore, the expressing genes will be the 4th row after another division, and switch to the 2nd row again after another. If the 3 patterns of gene expressions can be shifted in turn as expected, we can elongate the loop without a complicated gene design.
This is the exact same state as the phenotypic changes began, except that it is the genes in the 3rd row, instead of the 2nd, that are being expressed. In other words, the expressed genes are switched through one cell division. The gene groups are symmetric in terms of colors (red, blue and green) when the reagent does not exist. Therefore, the expressing genes will be the 4th row after another division, and switch to the 2nd row again after another. If the 3 patterns of gene expressions can be shifted in turn as expected, we can elongate the loop without a complicated gene design.
Above is the gene groups that we designed. Toehold switch takes a key role of the gene expression change, and makes it easy to expand the system because it has a lot of pairs which do not crosstalk (link to application & citation). Also, it is a translational regulator, not transcriptional, so the response is expected to be faster.
Sigma factors and anti-sigma factors enables the pattern of the gene expression to be looped among cell divisions.
Pnrd switches the gene expression at the timing of a cell division.In reality, there are problems such as the inconsistency of the timing when Pnrd works and cell division, (Pnrd actually works on the time of DNA replication, not that of cell division.) and the preference of sigma factors to bind with anti-sigma factors than sigma promoters(link to modeling, and experiment). However, various phenotypes can be realized theoretically according to our computer simulation, depending on the properties of the factors and promoters.
References
[1] Figure 1 (B), Green, A. A., Silver, P. A., Collins, J. J., & Yin, P. (2014). Toehold switches: de-novo-designed regulators of gene expression. Cell, 159(4), 925-939.
[2]2014 iGEM UT-Tokyo wiki : https://2014.igem.org/Team:UT-Tokyo/Counter/Project?page=Project-block&contents=Project-2
Result
Cell Cycle Dependent Promoter: Pnrd
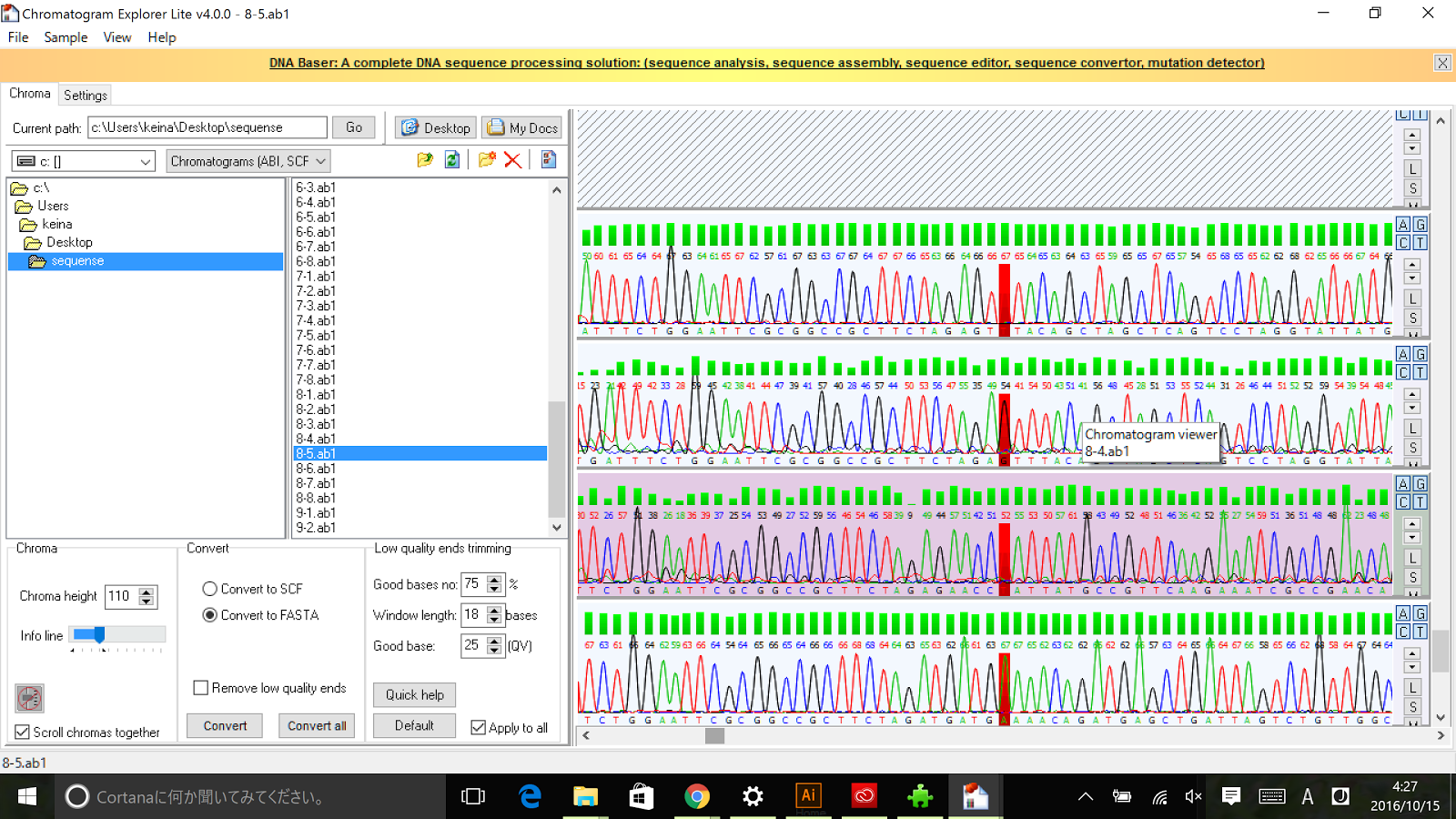
As described in the systems page, a cell division dependent promoter is essential for the functioning of our intergenerational phenotypic switching device. Stanford-Brown iGEM 2012 documented the characterisation of promoter Pnrd ([http://parts.igem.org/Part:BBa_K847211 BBa_K847211]) which activates during the cell division phase and confirmed its functionality. However, since it was submitted in RFC[25] standard, we decided to improve the part by using designing the part in RFC[10] in order to deal with the disadvantages of RFC[25] standard.
Sequencing results of Pnrd RFC[10]
The results of sequence show that the improved part contains the RFC[10] prefix and suffix.
With the RFC [10] standard, we characteri [[]]
Figure
sed the Pnrd promoter by ligating the promoter in front of part BBa_E0240 and compared it to the standard promoter BBa_J23101. The measurement was made approximately 4h after 100 fold dilution in M9 in a 5 min interval. For details of our characterisation, refer to our protocol. [[]]
FIgure
The results….
If positive result:
(briefly describe the graph, ……..) suggesting that the Pnrd was activated during cell division. Compared to BBa_J23101, the promoter is( …. ).
If negative results:
According to graph x, (....write something) showing insignificant difference in fluorescence with our negative control/ BBa_J23101. A possible explanation for the negative test results of the Pnrd promoter could be that the cells were not synchronised properly. Since we did not have DL-serine hydroxamate solution, we attempted to synchronise the cells using a stationary phase method. However, whether the cell were synchronised was not tested.
Application