Characterization of the K1372001 biobrick from the 2014 "This is not a lemon" Paris Saclay project
As a part of the characterization of a previous existing Biobrick Part, we chose the BBa_K13372001 biobrick from the Paris Saclay 2014 project “This is not a lemon”. It was designed to mimic the ripening of a lemon in E.Coli by the salicylate inducible expression a t-RNA suppressor.
The Paris Saclay 2014 team chose to use chromoproteins to express these colors in E. coli. Chromoproteins are reflective proteins that contain a pigmented prosthetic group and do not need to be excited to be seen. They fused a yellow chromoprotein with a blue one in order to display a green color. This construction is referred as the green fusion chromoprotein. In order to make the bacteria ripe like a real lemon, they decided to take advantage of the design of the fusion protein by using a translational suppression system. They added amber codons within the linker separating the yellow and the blue chromoproteins. Therefore, the expression of a suppressor t-RNA will suppress amber codons allowing the translation of the green fusion chromoprotein. Conversely, the down regulation of the suppressor t-RNA through time will allow bacteria switch from green to yellow, thus simulating the ripening of a real lemon. This system is referred to as the color switch system.
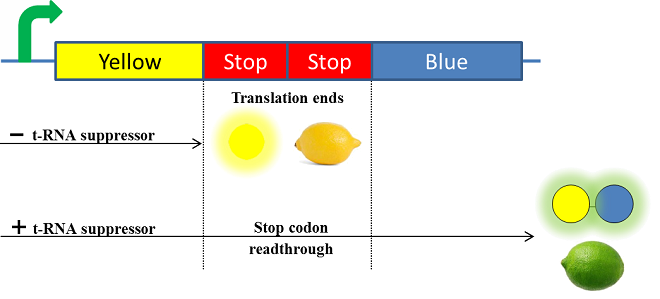
summary of the lemon ripening project
The t-RNA used is the supD suppressor t-RNA. It has been placed under control of a salicylate inducible promoter Psal to suppress the introduced amber codon. The nahR gene encodes a transcriptional regulator that is induced by salicylate and thus binds the nah or Psal promoters. In presence of a high salicylate concentration in the agar media supD will be expressed and so the green fusion chromoprotein: bacteria will display a green color. However, as bacteria grow into agar, less salicylate will remain available into the media. Thus, the decrease of the nahR-salicylate complex amount within bacteria will lead to supD downregulation through time. In turn, decrease of supD amount will result in less codon readthrough and so less translation of the green fusion protein and more translation of the yellow chromoprotein. As a result, bacteria will gradually change from green to yellow.
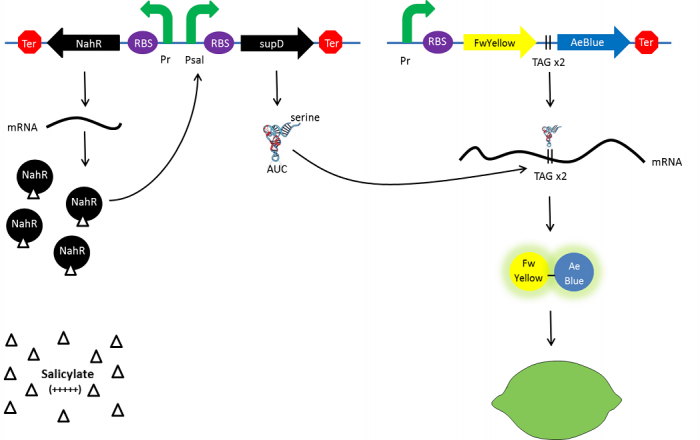
explanatory diagram of the lemon ripening
In order to characterize our biobrick, the color switch system (K1372001) was tested on three different green fusion chromoprotein constructions, as well as under three different salicylate concentrations. In order to achieve that, both measurements of Beta-Galactosidase (Fw Yellow) activity and of the luminescence resulting from Luciferase activity (Ae Blue) were made on bacterial cultures.
The experiment was conducted on three sets of cultures of bacteria containing three different green fusion chromoprotein constructions:
- “TAA”: BL21|K1372001 and pcl_TAA
- “Qt”: BL21|K1372001 and pcl_Tq
- “TAG”: BL21|K1372001 and pcl_TAG
and each of those sets of culture were incubated with three different salicylate concentrations : 0, 30µM and 1mM.
Each construction (TAA, Tq or TAG) was tested on 3 different clones (clones 1, 2 or 3), with 3 different salicylate concentrations (0, 30µM or 1mM), with in addition a negative control sample.
Pcl_TAA construction contains two TAA stop codon within the green fusion chromoprotein linker. This codon is not recognized by the supD suppressor t-RNA, thus no blue fluorescence is expected.
pcl_Tq construction does not contain any stop codon, thus the green fusion chromoprotein construction should entirely been transcripted and emit both yellow and blue fluorescence.
pcl_TAG contains the TAG codon recognized by supD suppressor t-RNA. Thus the transcription and the blue fluorescence of AeBlue should be inducible by salicylate.
The luciferase luminescence is expected to vary depending to the different constructions conditions and to salicylate concentrations, instead of the Beta Galactosidase activity which will remain constant. Thus luciferase data were normalized with those from Beta Galactosidase and our results are expressed as the Luciferase/Beta-Galactosidase activity.
In the Tq conditions, the plasmid does not contain any stop codon between the FwYellow’s open reading frame and the AeBlue’s open reading frame. Thus, no matter the salicylate concentration, the Luciferase activity is supposed to be highly detected, and the ratio luciferase/bGal should be superior to 1.
As expected a very high level of Luciferase/bGal activity is observed, but a decrease is noticed with the rise of salicylate. Indeed, both activity of Luciferase and bGal drop from 30µM of salicylate.
We can make the hypothesis that the salicylate has a direct or indirect influence on the metabolism of bacteria, and thus could reduce the synthesis of protein, whose luciferase and beta-galactosidase.
In order to read the next results, we will evaluate the levels of activity in TAA and TAG constructions in comparison to those in Tq, and express it in percentage.
In the TAA conditions, regardless of the salicylate concentration, there is no significant activity of Luciferase/Beta-Galactosidase.
Those results indicate that bacteria with part K1321001 and the TAA green protein fusion are unable to emit blue fluorescence due to the luciferase activity.
The stop codon TAA does not seem to be recognized by the supD tRNA suppressor, and
In TAG condition we can see a rising of the luciferase/bGal activity with the increase of the salicylate concentration.
In comparison to the results obtained with the TAA construction, the luciferase fluorescence activity is way higher for each salicylate condition. Thus this luminescence is specific to the TAG green protein fusion, and more precisely, to the TAG stop codon. This proves the good transcription and translation of the AeBlue gene, in consequence to a high TAG read-through in presence of supD.
The suppressor RNAt supD is specific to TAG codon, and cannot ensure the full transcription of the green protein fusion TAA.
In addition, the luciferase/bGal activity sharply rises from 30µM of salicylate in the TAG construction, but stays null for TAA. Thereby the variations of salicylate concentration are responsible of the increased production of supD specific to the TAA codon.

