(Add MFC constructs.) |
m (Add SBOL visual diagrams for MFC constructs.) |
||
| Line 656: | Line 656: | ||
<FIGURE> | <FIGURE> | ||
| − | <IMG SRC=""> | + | <IMG SRC="https://static.igem.org/mediawiki/2016/2/22/T--Newcastle--Parts-OmF_Expression_D.png"> |
<FIGCAPTION>SBOL Visual Diagram of our OprF expression construct.</FIGCAPTION> | <FIGCAPTION>SBOL Visual Diagram of our OprF expression construct.</FIGCAPTION> | ||
</FIGURE> | </FIGURE> | ||
| Line 700: | Line 700: | ||
<p> | <p> | ||
<FIGURE> | <FIGURE> | ||
| − | <IMG SRC=""> | + | <IMG SRC="https://static.igem.org/mediawiki/2016/0/0a/T--Newcastle--Parts-OprF_Expression_D.png"> |
<FIGCAPTION>SBOL Visual Diagram showing our OprF expression construct.</FIGCAPTION> | <FIGCAPTION>SBOL Visual Diagram showing our OprF expression construct.</FIGCAPTION> | ||
</FIGURE> | </FIGURE> | ||
Revision as of 14:44, 11 October 2016
Parts
You can also find all the parts we've designed in the parts registry. We've used SBOL visual to specify our designs.
Electrically Induced 'Light Bulb'
We plan to clone Escherichia coli so that it fluoresces when a current of around 400mA is passed through the broth that the E. coli are growing in.
Previous attempts at this experiment have used GFP to fluoresce with the current. In terms of this experiment we shall use GFP to ensure the design construct works. We have incorporated restriction sites within the construct so that we are able to then switch the GFP with other constructs, such as luciferase to emit light like an actual lightbulb.
In order for the GFP to be activated by a current, a heat-shock induced promoter must be present. Previous experiments have used sigma 32 as a heat shock promoter which seems to have the desired effect, although we shall test this in the lab. We have also inserted two restriction sites to allow us to either cut out the sigma 32 promoter, or add an extra promoter. This will allow us to test the effect that the sigma 32 has on the GFP production and whether it is induced by heat-shock or not.
The design has a natural ribosome binding site, which we will be adding to the registry (BBa_K1895001). This is to ensure that the ribosome does bind to the DNA and synthesise the protein correctly, however we will make variants of this DNA with two different medium bicistronic rbs. The medium bicistronic rbs will avoid the problem of placing too high a translational burden on the cell. We will then test all three variants to determine which is the best rbs to use in the final design.
Our Construct


Arabinose Controlled 'Variable Resistor'
We plan to engineer Escherichia coli to behave like a variable resistor. We aim to do this by using E. coli to vary the amount of free ions in an electrolyte. Ion uptake will be controlled by the expression of smtA. SmtA is a metallothionein that can bind to heavy metal ions like cadmium (II), Zinc (II) and Copper (II).
SmtA has been used in a number of iGEM projects and is in the registry (BBa_K519010). It has previously been used in experiments for Cadmium (II) uptake, see Tokyo-NokoGen 2011. We will be examining firstly, the impact of smtA of Zinc (II) concertation rather than Cadmium (II) and then the impact that this has on the resistivity of the Zinc (II) containing media. In this instance we will be using Zinc sulfate (ZnSO4) in solution where it disassociates into Zn2+ and SO42- ions. Various concentrations of Zinc sulfate have known electrical conductivity . When smtA is expressed it will render the Zn2+ unavailable and thereby reduce the conductivity of the solution.
We will be placing smtA under the control of an AraC regulated promoter allowing the expression of smtA to be controlled by the addition or removal of arabinose.
Our Construct
We will be synthesising two variants of our construct, one with and one without an ssRA degradation tag. This will allow us to see if we gain finer control over the resistance by increasing the rate of protein degradation. Both variants of the construct will include a polyhistidine-tag to allow for protein purification from cultures.
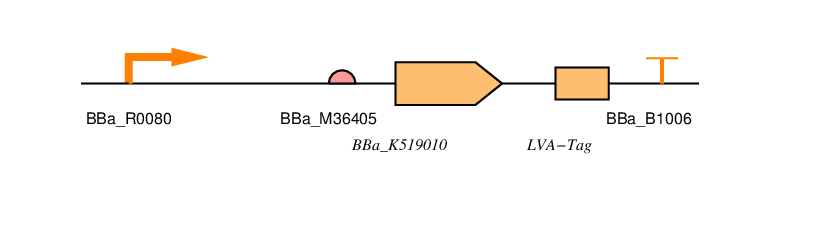
BBa_R0080
This is the AraC regulated promoter. The way this promoter behaves is that transcription takes place in the presence of AraC or arabinose. Without arabinose present there should be no transcription.
BBa_B0034
This is a standard RBS based that used in the construction of the repressilator. (Elowitz, 1999). It has an efficiency of 1. Whilst not the most efficient RBS it has a high efficiency and is widely used in iGEM projects. It is also present in the 2016 distribution kit.
BBa_K519010
This is the coding sequence for SmtA originally from Synechococcus sp, a cyanobacterial strain.
LVA-TAG
This is an ssRA protein degradation tag. Tagged proteins are degraded by the proteases ClpXP or ClpAP. There are a number of tag sequences, variants of AANDENYALAA, with the last three amino acids varying. The last three amino acids determine the half-life of the protein. LVA is a fast protein degradation tag. We use this to ensure that the resistance is reduced quickly after removal of arabinose.
BBa_B1006
This is an artificial terminator part and was chosen because it has a high forward efficiency of 0.99.
pSB1C3
We are using the standard BioBrick backbone part pSB1C3 as this will make it easier to submit the part to the registry at a later date.
We have included restriction sites around protein and tag so that it can be replaced with a part without the tag to see if this has any effect on Zinc uptake.
OmpR Controlled Red 'Light Dependent Resistor'
We plan to engineer Escherichia coli to behave like a light dependent resistor. We aim to do this by using E. coli to vary the amount of free ions in an electrolyte in response to light. Ion uptake will be controlled by the expression of smtA. SmtA is a metallothionein that can bind to heavy metal ions like cadmium (II), Zinc (II) and Copper (II).
SmtA has been used in a number of iGEM projects and is in the registry (BBa_K519010). It has previously been used in experiments for Cadmium (II) uptake, see Tokyo-NokoGen 2011 and for accumulating Zinc (II) intracellularly. We will be examining firstly, the impact of smtA on Zinc (II) concertation rather than Cadmium (II) and then the impact that this has on the resistivity of the Zinc (II) containing media. In this instance we will be using Zinc sulfate (ZnSO4) in solution where it disassociates into Zn2+ and SO42- ions. Various concentrations of Zinc sulfate have known Electrical conductivity . When smtA is expressed it will render the Zn2+ unavailable and thereby reduce the conductivity of the solution.
For this LDR we will be using the red light detection system from the Colliroid project ( paper). In this scheme the production of SmtA which affects the resistivity is placed under the control of the OmpF upstream promoter (BBa_R0084). We propose to engineer a system where this promoter is repressed in the dark and has increased transcription in (red) light. This allows the device to mimic the behaviour of a traditional electronic LDR whereby resistance is decreased in the light and increased in the dark.
The OmpF promoter is repressed by phosphorylated OmpR, OmpR-P. In normal conditions the E. coli cell contains free OmpR which can be phosphorylated by expression of a protein with an EnvZ domain. One such protein is the fusion protein, Cph8 (BBa_I15010). In the dark this protein phosphorylates OmpR and so prevents SmtA production, increasing resistance. In the light, the light responsive domain Cph1 inhibits the activity of the EnvZ is prevented from phosphorylating OmpR and therefore allows SmtA production and decreased resistance.
Note that this will only work in E. coli which are naturally deficient in EnvZ.
In order for the light responsive domain of the fusion protein cph8 to sense red light the formation of a chromophore is required this is done by the production of two proteins, ho1 and PcyA together with the cph8. In our system these will be constitutively expressed to create the red light sensor.
Our Construct
There are two parts to our construct, the red light sensing component and the SmtA production component. It is presented below as a one plasmid system with the following parts.
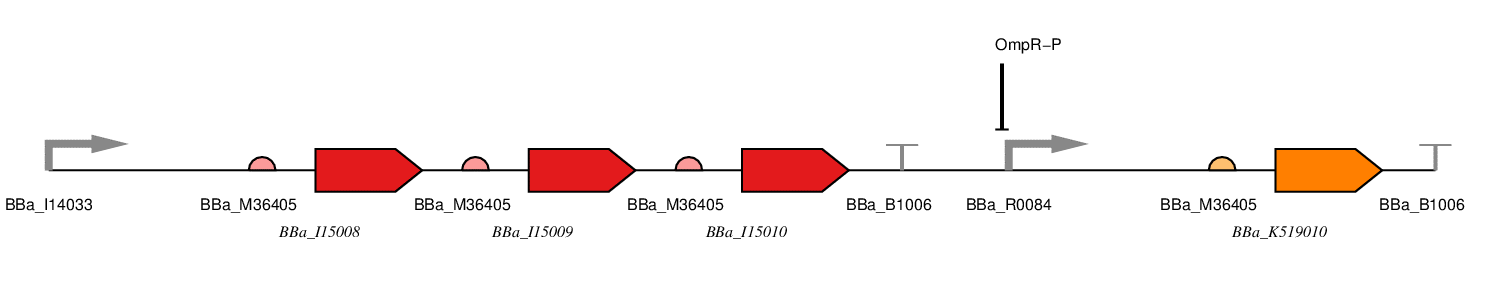
Figure 1: Red LDR.
BBa_J23100 - constitutive promoter
We will use a σ70 constitutive promoter as this is the main E. coli sigma factor. Consequently, there should be RNA polymerase present to transcribe from this promoter at all stages during the bacterial growth cycle. Specifically, we have chosen BBa_J23100, an artificial promoter due to its widespread use, documentation and comparatively short sequence (35bp).
BBa_I15008 - ho1
This is one of the proteins required for chromophore formation.
BBa_I15009 - PcyA
This is one of the proteins required for chromophore formation.
BBa_I15010 - cph8
This is a fusion protein consisting of a light receptor domain and EnvZ domain. In the dark the EnvZ domain of this protein phosphorylates the free OmpR in the cell which represses the OmpF promoter and induces the OmpC promoter.
BBa_R0084 - OmpR-P Promoter
This part is the promoter usually found upstream of OmpF. It is repressed by phosphorylated OmpR.
BBa_K519010 - SmtA
This is the coding sequence for SmtA originally from Synechococcus sp, a cyanobacterial strain.
BBa_B1006 - Terminator
This is an artificial terminator part and was chosen because it has a high forward efficiency of 0.99.
pSB1C3 - Backbone
We are using the standard BioBrick backbone part pSB1C3 as this will make it easier to submit the part to the registry at a later date.
Construction
BioBrick Assembly
The fusion protein cph8 (BBa_I15010) is not available in the distribution kit, it is however in stock at the registry and can be ordered. The remaining parts are in the distribution so BioBrick assembly can be performed. However because of the large number of parts, and there are no intermediaries in the registry or distribution kit, this construct would be unwieldy to assemble in this manner.
YF1-FixJ Controlled Blue 'Light Dependent Resistor'
We plan to engineer Escherichia coli to behave like a light dependent resistor. We aim to do this by using E. coli to vary the amount of free ions in an electrolyte in response to light. Ion uptake will be controlled by the expression of smtA. SmtA is a metallothionein that can bind to heavy metal ions like cadmium (II), Zinc (II) and Copper (II).
SmtA has been used in a number of iGEM projects and is in the registry BBa_K519010. It has previously been used in experiments for Cadmium (II) uptake, see Tokyo-NokoGen 2011 and for accumulating Zinc (II) intracellularly. We will be examining firstly, the impact of smtA of Zinc (II) concertation rather than Cadmium (II) and then the impact that this has on the resistivity of the Zinc (II) containing media. In this instance we will be using Zinc sulfate (ZnSO4) in solution where it disassociates into Zn2+ and SO42- ions. Various concentrations of Zinc sulfate have known electrical conductivity . When smtA is expressed it will render the Zn2+ unavailable and thereby reduce the conductivity of the solution.
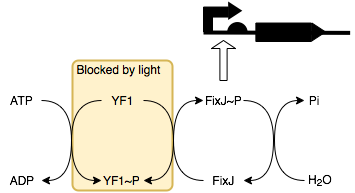 |
We will be placing smtA under the control of a FixJ-P (phosphorylated FixJ)
promoter. This allows it to be regulated by blue light through a series of
reactions with its response regulator protein YF1 (below).
In the absence of light, YF1 undergoes autophosphorylation to produce YF1-P which can then phosphorylate FixJ. This in turn activates the transcription of the downstream protein, in this case it is SmtA. Thus, in the presence of light SmtA is not produced and so conductivity does not change, whilst in the absence of light SmtA is produced resulting in a decrease in resistance.
Clearly, this behaviour is the inverse of an electrical light dependent resistor where resistance increases with light intensity. To mimic this behaviour using biological circuits we would place an inverter before the FixK2 promoter (which is activated by FixJ-P). The inverter is constructed by placing the desired output, here SmtA, under the control of a lambda cl regulated promoter (BBa_R0051). As lambda cl represses the promoter having this produced under control of FixK2 promoter inverts the system so that SmtA is produced in the presence of light rather than the absence thereof. BBa_K592020 is an example of a part that uses this technique.
Our Construct
The non-inverted construct is shown in Figure 1 and the inverted construct in Figure 2. Currently our device is shown as a 1 plasmid system but there is no reason that the two separate sub-components could not be split into a 2 plasmid system for easier assembly.

Figure 1: Standard Blue LDR construct.

Figure 2: Blue LDR with inverter.
For the non-inverted construct the parts are as follows:
BBa_J23100 - constitutive promoter
We will use a σ70 constitutive promoter as this is the main E. coli sigma factor. Consequently, there should be RNA polymerase present to transcribe from this promoter at all stages during the bacterial growth cycle. Specifically, we have chosen BBa_J23100, an artificial promoter due to its widespread use, documentation and comparatively short sequence (35bp).
BBa_K592016 - FixJ & YF1 with RBSs
The YF1 and FixJ coding sequences are provided as a composite part together with standard RBSs in part BBa_K592016 which we have chosen for ease of assembly, in the event that we or future teams wish to use the BioBrick standard assembly to produce our part.
BBa_K592006 - FixK2
This is the wild-type promoter to which phosphorylated FixJ binds. It is reported that this promoter has very little leaky activity in the absence of FixJ.
BBa_K519010 - SmtA
This is the coding sequence for SmtA originally from Synechococcus sp, a cyanobacterial strain.
BBa_B1006 - Terminator
This is an artificial terminator part and was chosen because it has a high forward efficiency of 0.99.
pSB1C3 - Backbone
We are using the standard BioBrick backbone part pSB1C3 as this will make it easier to submit the part to the registry at a later date.
For the inverted part there are additional parts as follows:
BBa_C0051 - Lambda CI
This is the repressor protein from Lambda phage it represses the promoter BBa_R0051.
BBa_B0010 and BBa_B0012 - Double stop terminators
These are the terminators used in the composite part BBa_S04617 which is replicated in our construct.
BBa_R0051 - Lambda CI controlled promoter
This is a promoter from Lambda phage that is repressed by lambda Cl (BBa_C0051).
Construction
Synthesis
This construct can be sourced from IDT using our free allowance.
BioBrick Assembly
There exist a number of intermediate assembly components in the parts distribution that can be used to assemble our part faster if we use BioBrick assembly. Notably, BBa_S04617 contains the inverter,BBa_K592016 contains the FixJ and YF1. The two devices can be constructed separately as follows.
Constitutive Production Device
-
Cut the terminator BBa_B1006 with E & X.
-
Cut BBa_K592016 with E & S.
-
Mix & Ligate to form intermediate YF1FixJ+Terminator.
-
Cut the intermediate part with E & X and the constitutive promoter BBa_J23100 with E & S.
-
Mix and Ligate to form the constitutive production device.
SmtA Expression Device (inverted)
-
Cut BBa_K519010 with E & X.
-
Cut BBa_S04617 with E & S.
-
Mix and ligate to produce intermediate part: inverted smtA production.
-
Cut terminator with E & X.
-
Cut SmtA production intermediate with E & S.
-
Mix and ligate to produce SmtA expression device.
To produce the non-inverted device replace BBa_S04617 with the SmtA coding sequence and use an additional step to join this to an RBS, we suggest the standard RBS BBa_B0034 as this has good efficiency.
Biological 'Capacitor'
We plan to engineer Escherichia coli to mimic one of the properties of a capacitor, the ability to accumulate and hold charge for some time before discharging. This is shown in the idealised graph below.

An electrical capacitor accumulates charge whilst a voltage is applied and then discharges when the voltage stops being applied. We make an analogy between the voltage signal and protein concentration. Whereas an electrical capacitor accumulates charge, a biological ‘capacitor’ would accumulate proteins. Like an electrical capacitor which has a maximum charge it can accumulate, there is a maximum protein concentration that can accumulate in the cell determined by its production and degradation rate. Once proteins stop being accumulated in the cell it ‘discharges’ by having these drive the production of an output signal.
In this construct we use L-arabinose to mimic a voltage signal. This is entirely for experimental purposes, there is no reason that this device cannot be modified to respond to an electrical signal, for instance through the heat shock response explored elsewhere in our work. Although in this case we show that protein’s can be accumulated there is no reason why actual charge, in the form of a potential difference could not be generated across the cell membrane. There are already examples of membrane potentials in biology, the most obvious being found in neurons. This is something that has been explored by iGEM teams in the past e.g., Cambridge (2008). Importantly we show through modelling that the charge-discharge cycle can be mimicked in biological cells through the use of repressor/inducer competition. This could be merged with work on membrane potentials in the future.
Because we use a constitutively on promoter, the TetR repressible promoter (BBa_R0040) the default state of the system is ‘charging’. In this state lambda repressor (BBa_C1051) accumulates in the cell together with 434 repressor (BBa_C0052). The amount of 434 repressor grows faster than that of lambda repressor because there are two coding sequences for the protein in the circiut. This is to ensure that it outcompetes (on average) the lambda repressor so that there is a low output signal whilst in the charging state. This occurs because 434 repressor represses the output promoter whilst lambda repressor induces it. In out device the output signal is sfGFP (BBa_I746916).
We can switch the state of the system to the discharging state by causing the expression of TetR. To facilitate this we have used an L-arabinose promoter coupled with the TetR coding sequence to give us a chemical ‘off switch’. Once the TetR is produced the system enters the discharging state, no further protein synthesis in our construct is induced and so the amount of 434 repressor and lambda repressor start to decay. The 434 repressor is tagged with a very fast ssRA degradation tag, the LVA degradation tag so that it will be broken down faster than lambda repressor.
As this happens there will reach a point where the 434 repressor stops out-competing the lambda repressor and the output will start to be produced as it is induced by the lambda repressor. Whilst this is happening the level of 434 and lambda repressor will continue to fall until the output stops being produced and the system has completely ‘discharged’ and is in a resting state. At this point the removal of L-arabinose and the addition of tetracycline or an analogue thereof (which binds to TetR and prevents it from repressing the promoter) would switch the system back into the charging state and the process can begin again.
Construct
Our construct, as shown here, is a composite part which is built entirely of BioBricks already in the registry. The parts used are as follows.
| Part No. | Name | Purpose |
|---|---|---|
| BBa_R0080 | L-Arabinose Promoter | This part is an L-Arabinose inducible promoter with very low level expression in the absence of L-Arabinose and AraC. Be aware, if the E. coli strain used constitutively expresses AraC then this promoter will ‘leak’. Check the strain list for information. |
| BBa_R0040 | TetR | This is the coding sequence for TetR which represses BBa_R0040. |
| BBa_R0040 | TetR Repressible Promoter | This is a constitutively on promoter which can be repressed by TetR. Be aware, if the strain used expresses TetR constitutively then this promoter will be repressed. Check the strain list for information. |
| BBa_C0052 | 434 Repressor | Represses the output promoter. |
| BBa_C0051 | Lambda Repressor | Induces the output promoter. |
| BBa_I12006 | Modified Promoter Part | This is a modified promoter part, originally the lambda Prm promoter. The modification allows it to be activated by lambda repressor and repressed by 434 repressor. |
| BBa_I746916 | Superfolder GFP | This is the coding sequence for super folder GFP. We have chosen to use this as our reporter because it can easily be quantified using a plate by taking the OD600 measurement. This is harder to quantify with more visible reporters like amilCP. |
| BBa_B1006 | Standard Terminator | We chose to use this promoter from the registry as it has a high forward efficiency. |
| BBa_B0034 | RBS | We chose to use this RBS from the registry as it is efficient and widely used in iGEM projects. |
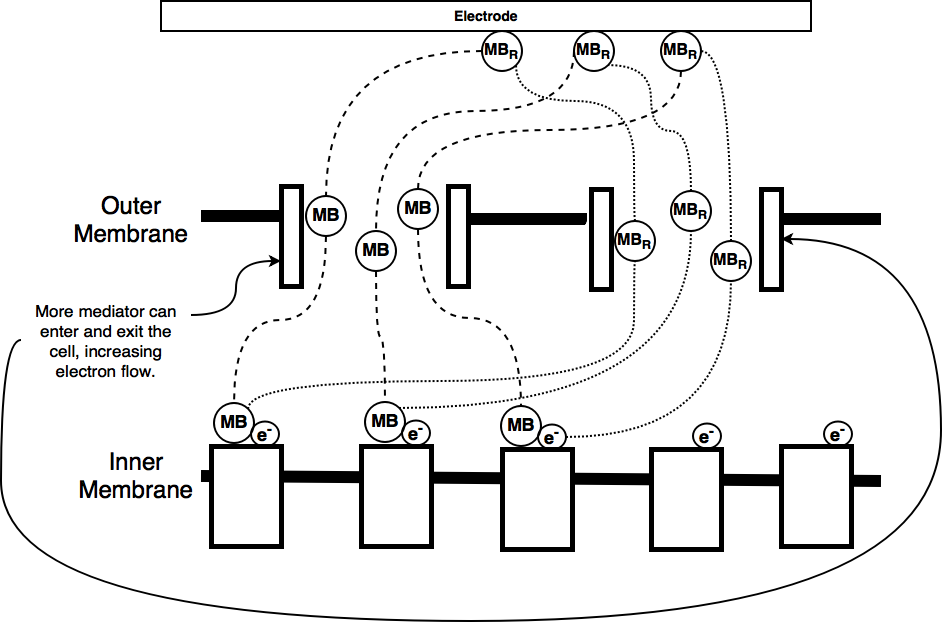
As part of our iGEM project we worked on trying to improve the current output from a microbial fuel cell. Microbial fuel cells which use E. coli depend on a mediator, usually methylene blue to transfer electrons between the electrodes and the organisms electron transport chain. The mediator enters the cell through porins in the cell membrane and 'steals' electrons, becoming oxidised. These electrons are then carried to the cathode, which is positively charged where they are lost, allowing electrons to flow around the circuit.
By increasing the amount of mediator that can enter the cell we hope to be able to increase the electron flow rate, or current, from our microbial fuel cell. This occurs because the electrons can be moved more freely and they're movement via the mediator is less limited by the rate at which the cell can transport the mediator across the membrane.
One of the ways of increasing the amount of mediator entering and exiting the cell is to increase the number of porins. For this purpose we have designed this device to control the overexpression of one of the E. coli natural porins, OmpF.
We chose OmpF because it occurs naturally in E. coli and is suitable for the mediator we use in our fuel cell (which is a variant of the University of Reading's NCBE kit). OmpF is permeable for molecules smaller than 600 Da whilst methylene blue is 284 Da.
By adding L-arabinose to the grown media the number of OmpF porins in the E. eoli membrane should increase, allowing increased extracellular transport of methylene blue and faster electron shuttling between the cells and the electrodes. This should correspond to an increase in current. This function is shown below.

Our OmpF overexpression device is simple in construction, consisting of a pBad promoter, standard RBS, the OmpF sequence and a standard terminator. This is part number BBa_K1895004 in the registry.

| Part No. | Name | Purpose |
|---|---|---|
| BBa_K206000 | pBAD Strong | Promotes the downstream expression of OmpF when L-arabinose is present in the growth medium. |
| BBa_B0034 | RBS (Elowitz) | A standard, efficient RBS, as used in our other constructs. |
| BBa_K864204 | OmpF | This is the coding sequence for OmpF, a natural porin protein found in E. coli. and involved in the transport of small molecules across the cell membrane. |
| BBa_B0012 | Terminator | This is a standard, widely used terminator in iGEM projects. We switched to using this terminator when we had synthesis problems with the double loop terminator used in our other constructs (BBa_B1006). |
We are also investigating an alternative way of increasing the amount of mediator transport through our work which is the expression of a larger membrane porin. One of the limiting factors of a microbial fuel cell is the low permeability of the cell membrane which limits mediator transport. Team Bielefeld in 2013 expressed the OprF porin protein from Pseudomonas fluorescens in E.coli which increased cell membrane permeability. This is because the porin size is much larger than E. coli's natural porins (such as OmpF, above). This allows more mediator to pass through the membrane.
One of the problems the Bielefeld team reported was the reduced growth of the E. coli due to metabolic stress. This is particularly noticeable when using the T7 promoter, as in their part BBa_K1172502. Population size is important for fuel cell output, the more cells there are the more electron donors there are and so theoretically the greater the current. To overcome this issue we have changed their promoter to a pBAD promoter which is regulated by L-arabinose monosaccharide. Thus replication stress is reduced during the first part of the growth curve. This should allow the bacterial population to grow rapidly in the fuel cell, ensuring we get a large population before L-arabinose is added (this increased level of growth should be seen by a higher OD600 value closer to 4.0 (growth of wild type) after 10 hours of growth). At this point the current should then further increase due to the larger porin expression.
This part is similar in construction to our OmpF expression part, with the protein coding sequence swapped to OprF rather than OmpR. This is part number BBa_K1895005 in the registry.

| Part No. | Name | Purpose |
|---|---|---|
| BBa_K206000 | pBAD Strong | Promotes the downstream expression of OmpF when L-arabinose is present in the growth medium. |
| BBa_B0034 | RBS (Elowitz) | A standard, efficient RBS, as used in our other constructs. |
| BBa_K1172501 | OmpF | This is the coding sequence for OrpF, a large porin protein found in Pseudomonas fluorescens. |
| BBa_B0012 | Terminator | This is a standard, widely used terminator in iGEM projects. We switched to using this terminator when we had synthesis problems with the double loop terminator used in our other constructs (BBa_B1006). |

