Gracetexana (Talk | contribs) |
Gracetexana (Talk | contribs) |
||
| Line 10: | Line 10: | ||
<h2>Registry Parts</h2> | <h2>Registry Parts</h2> | ||
<br> | <br> | ||
| + | <div class="column half_size"> | ||
<h3>pH Sensors</h3> | <h3>pH Sensors</h3> | ||
| − | < | + | </html> |
| + | [[File:T--Austin_UTexas--Cpx_pH_Culture_Tubes_2.png|thumb|left|300px| Figure 1. Testing the CpxR Construct in pH 6-9. From left to right is Control pH 6-9 and then Experimental pH 6-9. These are showing the gradient change in expression accordingly with the change of pH due to a pH-dependent promotor compared to consistent expression accordingly with a promoter that is always "on".]] | ||
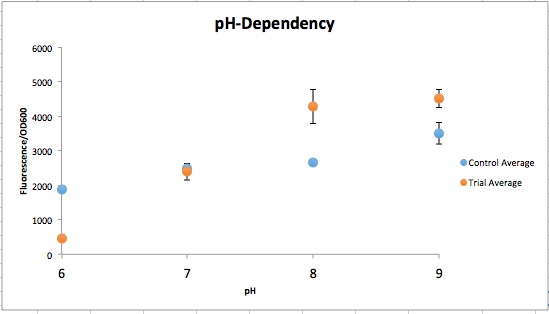
| + | [[File:T--Austin_UTexas--pH_Dependent_Promoter.jpeg|thumb|left|500px| Figure 2. Normalized fluorescent values from CpxR construct vs control (YGCP). The fluorescence per cell count stayed generally the same throughout the range of pH while the CpxR has a clear increase in fluorescence per cell.]] | ||
| + | |||
| + | <html> | ||
<h4>CpxA-CpxR</h4> | <h4>CpxA-CpxR</h4> | ||
<p> | <p> | ||
CpxA-CpxR is a two-component mechanism that is activated at pH 7.4 and repressed at pH 6.0. CpxA is an intermembrane protein that autophosphorylates at a certain external pH, CpxR (a kinase) then gets phosphorylated by CpxA and acts as a transcription factor. This system originally is a transcription factor for the virF gene, but we replaced virF with the Reporter. The original sequence was found in <i>Shigella sonnei</i>, but E. coli has a homolog of these proteins so all that is required on the construct is the appropriate prefix/suffix and CpxR binding site. | CpxA-CpxR is a two-component mechanism that is activated at pH 7.4 and repressed at pH 6.0. CpxA is an intermembrane protein that autophosphorylates at a certain external pH, CpxR (a kinase) then gets phosphorylated by CpxA and acts as a transcription factor. This system originally is a transcription factor for the virF gene, but we replaced virF with the Reporter. The original sequence was found in <i>Shigella sonnei</i>, but E. coli has a homolog of these proteins so all that is required on the construct is the appropriate prefix/suffix and CpxR binding site. | ||
<p> | <p> | ||
| − | |||
| − | |||
| − | |||
<p> | <p> | ||
Figure 1 qualitatively depicts the Control at pH 6 with more expression of the Yellow-Green Chromoprotein than the Experimental at pH 6. The pH-Dependent promoter of the Experimental group is down-regulated at pH 6 whereas the control is not. Also, there is an increase in YGCP expression between the Experiment pH 7 and pH 8 that is not seen in the Control between pH 7 and pH 8. The normalized data in Figure 2 shows the relative expression of YGCP since the qualitative data is ambiguous. The construct can be found on the iGEM registry as: <a href="http://parts.igem.org/Part:BBa_K2097000">K2097001</a>. | Figure 1 qualitatively depicts the Control at pH 6 with more expression of the Yellow-Green Chromoprotein than the Experimental at pH 6. The pH-Dependent promoter of the Experimental group is down-regulated at pH 6 whereas the control is not. Also, there is an increase in YGCP expression between the Experiment pH 7 and pH 8 that is not seen in the Control between pH 7 and pH 8. The normalized data in Figure 2 shows the relative expression of YGCP since the qualitative data is ambiguous. The construct can be found on the iGEM registry as: <a href="http://parts.igem.org/Part:BBa_K2097000">K2097001</a>. | ||
<p> | <p> | ||
| − | </ | + | </div> |
| − | + | <div class="column half_size" > | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <div class="column | + | |
<h3>Composite Parts</h3> | <h3>Composite Parts</h3> | ||
</html> | </html> | ||
| − | [[File:T--Austin_UTexas--BCPplate.png|thumb|left| | + | [[File:T--Austin_UTexas--BCPplate.png|thumb|left|500px| Figure 3. Control amilCP. This control was used to compare the color intensity to the pH sensitive P-atp2 while testing with a range of pH values. This part was documented to not be affected by changes in pH.]] |
<html> | <html> | ||
| Line 38: | Line 36: | ||
<p> | <p> | ||
<a href="http://parts.igem.org/Part:BBa_K2097001">K2097001</a> is a composite part made up of BBa_K592009 (blue chromoprotein) and BBa_K608002 (a strong RBS and promoter). The protein exhibits a strong blue/purple color when expressed. BBa_K608002 is made up of a promoter, BBa_J23104, and an RBS, BBa_B003. <b>LINKS</b> | <a href="http://parts.igem.org/Part:BBa_K2097001">K2097001</a> is a composite part made up of BBa_K592009 (blue chromoprotein) and BBa_K608002 (a strong RBS and promoter). The protein exhibits a strong blue/purple color when expressed. BBa_K608002 is made up of a promoter, BBa_J23104, and an RBS, BBa_B003. <b>LINKS</b> | ||
| − | < | + | <h4>Yellow-Green Chromoprotein</h4> |
| − | + | ||
| − | + | ||
| − | + | ||
<p> | <p> | ||
<a href="http://parts.igem.org/Part:BBa_K2097002">K2097002</a> is a composite part made up of BBa_K1033916 (amajLime, a yellow-green chromoprotein)and BBa_K608006 (a medium promoter and RBS). <b>LINKS</b> | <a href="http://parts.igem.org/Part:BBa_K2097002">K2097002</a> is a composite part made up of BBa_K1033916 (amajLime, a yellow-green chromoprotein)and BBa_K608006 (a medium promoter and RBS). <b>LINKS</b> | ||
Revision as of 21:36, 17 October 2016
Austin_UTexas
Registry Parts
pH Sensors
CpxA-CpxR
CpxA-CpxR is a two-component mechanism that is activated at pH 7.4 and repressed at pH 6.0. CpxA is an intermembrane protein that autophosphorylates at a certain external pH, CpxR (a kinase) then gets phosphorylated by CpxA and acts as a transcription factor. This system originally is a transcription factor for the virF gene, but we replaced virF with the Reporter. The original sequence was found in Shigella sonnei, but E. coli has a homolog of these proteins so all that is required on the construct is the appropriate prefix/suffix and CpxR binding site.
Figure 1 qualitatively depicts the Control at pH 6 with more expression of the Yellow-Green Chromoprotein than the Experimental at pH 6. The pH-Dependent promoter of the Experimental group is down-regulated at pH 6 whereas the control is not. Also, there is an increase in YGCP expression between the Experiment pH 7 and pH 8 that is not seen in the Control between pH 7 and pH 8. The normalized data in Figure 2 shows the relative expression of YGCP since the qualitative data is ambiguous. The construct can be found on the iGEM registry as: K2097001.
Composite Parts
Blue Chromoprotein
K2097001 is a composite part made up of BBa_K592009 (blue chromoprotein) and BBa_K608002 (a strong RBS and promoter). The protein exhibits a strong blue/purple color when expressed. BBa_K608002 is made up of a promoter, BBa_J23104, and an RBS, BBa_B003. LINKS
Yellow-Green Chromoprotein
K2097002 is a composite part made up of BBa_K1033916 (amajLime, a yellow-green chromoprotein)and BBa_K608006 (a medium promoter and RBS). LINKS