Ianoverman (Talk | contribs) |
Ianoverman (Talk | contribs) |
||
| Line 218: | Line 218: | ||
<div class="naviSection" id="section6"> | <div class="naviSection" id="section6"> | ||
<h2>pH Sensors</h2> | <h2>pH Sensors</h2> | ||
| − | <p> | + | <p>Possessing the ability to monitor the brewing process of kombucha without disturbing the microenvironment and using a very visible color reporter would allow for greater insights as to how the populations of organisms and pH may change due to competition amongst other bacteria and yeast in the beverage and SCOBY of the kombucha. The byproducts produced by the kombucha as it brews causes the tea to become more acidic, leading to our team searching for pH sensitive promoters, and for ways to implement these into kombucha.</p> |
| − | Possessing the ability to monitor the brewing process of kombucha without disturbing the microenvironment and using a very visible color reporter would allow for greater insights as to how the populations of organisms and pH may change due to competition amongst other bacteria and yeast in the beverage and SCOBY of the kombucha. The byproducts produced by the kombucha as it brews causes the tea to become more acidic, leading to our team searching for pH sensitive promoters, and for ways to implement these into kombucha. | + | |
| − | <p> | + | <p>Though an acidic sensor was what was required for our kombucha analysis, the identification of sensors in other areas of the pH spectrum were explored as well. Three sequences were identified, the CadC operon for the acidic range, CpxA-CpxR complex for the neutral range, and the P-atp2 promoter from the BioBrick Registry (<a href="http://parts.igem.org/Part:BBa_K1675021">BBa_K1675021</a>) for the basic range. Each sequence was paired with a unique corresponding reporter sequence so that if each pH sensitive plasmid were in the same environment, the specific pH of the system could be seen. The reporters used were, <a href="http://parts.igem.org/Part:BBa_E1010">BBa_E1010</a> for the CadC construct, <a href="http://parts.igem.org/Part:BBa_K1033916">BBa_K1033916</a> for the CpxA-CpxR complex, and <a href="http://partsregistry.org/Part:BBa_K592009">BBa_K592009</a> for the P-atp2 promoter.</p> |
| − | Though an acidic sensor was what was required for our kombucha analysis, the identification of sensors in other areas of the pH spectrum were explored as well. Three sequences were identified, the CadC operon for the acidic range, CpxA-CpxR complex for the neutral range, and the P-atp2 promoter from the BioBrick Registry (<a href="http://parts.igem.org/Part:BBa_K1675021">BBa_K1675021</a>) for the basic range. Each sequence was paired with a unique corresponding reporter sequence so that if each pH sensitive plasmid were in the same environment, the specific pH of the system could be seen. The reporters used were, <a href="http://parts.igem.org/Part:BBa_E1010">BBa_E1010</a> for the CadC construct, <a href="http://parts.igem.org/Part:BBa_K1033916">BBa_K1033916</a> for the CpxA-CpxR complex, and <a href="http://partsregistry.org/Part:BBa_K592009">BBa_K592009</a> for the P-atp2 promoter. | + | |
| − | <p> | + | <p><u>CadC</u></p> |
| − | <u>CadC</u> | + | |
| − | <p> | + | <p>The CadC operon is a native pathway in <i>E. coli</i>, involved in the cadaverine synthesis pathway. The protein CadC protein on the operon is produced and activates segments downstream of the operon on the CadBA receptors. The CadC protein is pH sensitive to an external pH 5.5 and below, as well as lysine dependent. A point mutation on codon 265, in which argenine is converted to cystine, causes the CadC protein to become lysine independent.<sup>1</sup> </p> |
| − | The CadC operon is a native pathway in <i>E. coli</i>, involved in the cadaverine synthesis pathway. The protein CadC protein on the operon is produced and activates segments downstream of the operon on the CadBA receptors. The CadC protein is pH sensitive to an external pH 5.5 and below, as well as lysine dependent. A point mutation on codon 265, in which argenine is converted to cystine, causes the CadC protein to become lysine independent | + | <p>Unfortunately, we have been unable to grow the modified CadC operon in <i>E. coli</i> suggesting some form of cell toxicity. Due to this apparent toxicity, no data regarding this mutant CadC could be collected. Alternative candidates are being explored for other pH sensors that sense in the acidic range.</p> |
| − | <p> | + | |
| − | Unfortunately, we have been unable to grow the modified CadC operon in <i>E. coli</i> suggesting some form of cell toxicity. Due to this apparent toxicity, no data regarding this mutant CadC could be collected. Alternative candidates are being explored for other pH sensors that sense in the acidic range. | + | <p><u>CpxA-CpxR</u></p> |
| − | <p> | + | |
| − | <u>CpxA-CpxR</u> | + | <p>CpxA-CpxR is a two-component mechanism that is activated at pH 7.4 and repressed at pH 6.0. CpxA is an intermembrane protein that autophosphorylates at a certain external pH, CpxR (a kinase) then gets phosphorylated by CpxA and acts as a transcription factor. This system originally is a transcription factor for the virF gene, but virF was replaced with a reporter. The original sequence was found in <i>Shigella sonnei</i>, but E. coli has a homolog of these proteins so all that is required on the construct is the appropriate prefix/suffix and CpxR binding site.</p> |
| − | <p> | + | |
| − | CpxA-CpxR is a two-component mechanism that is activated at pH 7.4 and repressed at pH 6.0. CpxA is an intermembrane protein that autophosphorylates at a certain external pH, CpxR (a kinase) then gets phosphorylated by CpxA and acts as a transcription factor. This system originally is a transcription factor for the virF gene, but | + | |
| − | <p> | + | |
</html> | </html> | ||
| + | |||
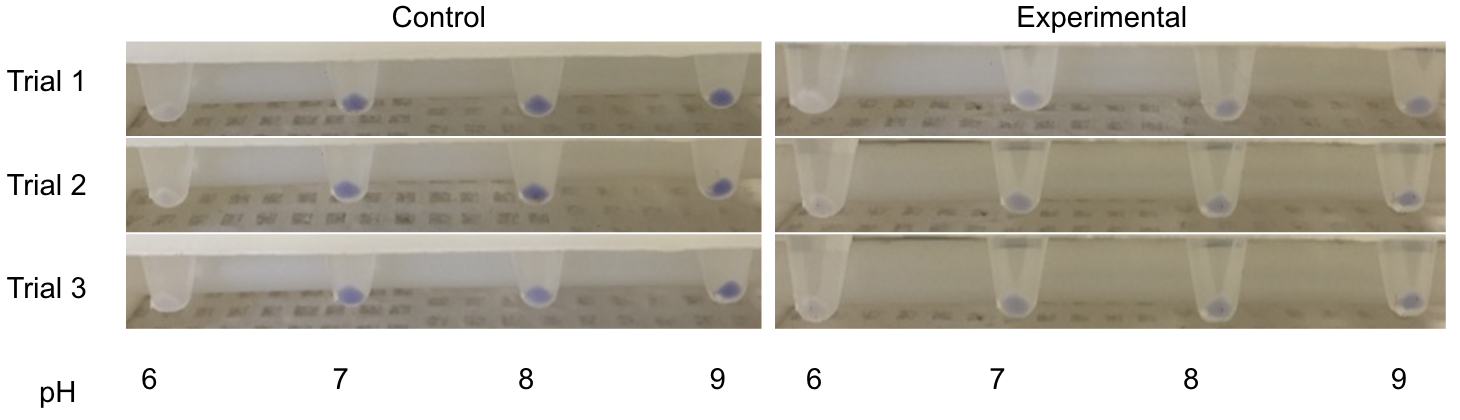
[[File:T--Austin_UTexas--Cpx_pH_Culture_Tubes_2.png|thumb|left|600px| Figure 1. Testing the CpxR Construct in pH 6-9. From left to right is Control pH 6-9 and then Experimental pH 6-9. These are showing the gradient change in expression accordingly with the change of pH due to a pH-dependent promotor compared to consistent expression accordingly with a promoter that is always "on".]] | [[File:T--Austin_UTexas--Cpx_pH_Culture_Tubes_2.png|thumb|left|600px| Figure 1. Testing the CpxR Construct in pH 6-9. From left to right is Control pH 6-9 and then Experimental pH 6-9. These are showing the gradient change in expression accordingly with the change of pH due to a pH-dependent promotor compared to consistent expression accordingly with a promoter that is always "on".]] | ||
<html> | <html> | ||
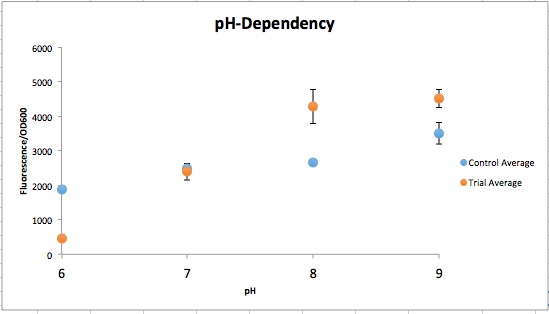
| − | <p> | + | <p>The order from left to right is control pH 6-9 and then Experimental pH 6-9. These are showing the gradient change in expression accordingly with the change of pH due to a pH-dependent promotor compared to consistent expression accordingly with a promoter that is always "on". The main point is that the Control at pH 6 has more expression of the yellow-green chromoprotein than the Experimental at pH 6. The pH-Dependent promoter of the Experimental group is down-regulated at pH 6 whereas the control is not. Also, there is an increase in YGCP expression between the Experiment pH 7 and pH 8 that is not seen in the Control between pH 7 and pH 8. The normalized data is below shows the relative expression of YGCP. The construct can be found on the iGEM registry as: <a href=“http://parts.igem.org/Part:Bba_K2097000”>BBa_K2097000</a>.</p> |
| − | The order from left to right is | + | |
| − | <p> | + | |
</html> | </html> | ||
| + | |||
[[File:T--Austin_UTexas--pH_Dependent_Promoter.jpeg|thumb|left|600px| Figure 2. Normalized fluorescent values from CpxR construct vs control (YGCP). The fluorescence per cell count stayed generally the same throughout the range of pH while the CpxR has a clear increase in fluorescence per cell.]] | [[File:T--Austin_UTexas--pH_Dependent_Promoter.jpeg|thumb|left|600px| Figure 2. Normalized fluorescent values from CpxR construct vs control (YGCP). The fluorescence per cell count stayed generally the same throughout the range of pH while the CpxR has a clear increase in fluorescence per cell.]] | ||
| Line 260: | Line 257: | ||
<ol type="1"> | <ol type="1"> | ||
<li>Hanke, T., Richhardt, J., Polen, T., Sahm, H., Bringer, S., and Bott, M. (2012) Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. <i>Journal of Biotechnology 157</i>, 359–372.</li> | <li>Hanke, T., Richhardt, J., Polen, T., Sahm, H., Bringer, S., and Bott, M. (2012) Influence of oxygen limitation, absence of the cytochrome bc1 complex and low pH on global gene expression in Gluconobacter oxydans 621H using DNA microarray technology. <i>Journal of Biotechnology 157</i>, 359–372.</li> | ||
| + | <li>CL, Dell., MN, Neely., and ER, Olson. (1994) Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. <i>Molecular Microbiology 14</i>, 7–16.</li> | ||
</ol> | </ol> | ||
</div> | </div> | ||
</html> | </html> | ||
Revision as of 06:26, 18 October 2016